Section 5 Adversarial computational control
library(h2o)
library(MLmetrics)
library(mltools)
library(scales)
library(enrichvs)
library(rlist)
library(stringr)
library(digest)
library(DT)
library(reshape2)
library(ggplot2)
library(ggforce)
library(reticulate)
use_condaenv("/Users/renee/Library/r-miniconda/envs/peptide_engineering/bin/python")5.1 General code/functions
# h2o.init(nthreads=-1, max_mem_size='100G', port=54321)
h2o.init(nthreads = -1)
h2o.removeAll()
# DeepLearning Grid 1
deeplearning_params_1 <- list(
activation = "RectifierWithDropout",
epochs = 10000, # early stopping
epsilon = c(1e-6, 1e-7, 1e-8, 1e-9),
hidden = list(c(50), c(200), c(500)),
hidden_dropout_ratios = list(c(0.1), c(0.2), c(0.3), c(0.4), c(0.5)),
input_dropout_ratio = c(0, 0.05, 0.1, 0.15, 0.2),
rho = c(0.9, 0.95, 0.99)
)
# DeepLearning Grid 2
deeplearning_params_2 <- list(
activation = "RectifierWithDropout",
epochs = 10000, # early stopping
epsilon = c(1e-6, 1e-7, 1e-8, 1e-9),
hidden = list(c(50, 50), c(200, 200), c(500, 500)),
hidden_dropout_ratios = list(
c(0.1, 0.1), c(0.2, 0.2), c(0.3, 0.3),
c(0.4, 0.4), c(0.5, 0.5)
),
input_dropout_ratio = c(0, 0.05, 0.1, 0.15, 0.2),
rho = c(0.9, 0.95, 0.99)
)
# DeepLearning Grid 3
deeplearning_params_3 <- list(
activation = "RectifierWithDropout",
epochs = 10000, # early stopping
epsilon = c(1e-6, 1e-7, 1e-8, 1e-9),
hidden = list(c(50, 50, 50), c(200, 200, 200), c(500, 500, 500)),
hidden_dropout_ratios = list(
c(0.1, 0.1, 0.1), c(0.2, 0.2, 0.2),
c(0.3, 0.3, 0.3), c(0.4, 0.4, 0.4),
c(0.5, 0.5, 0.5)
),
input_dropout_ratio = c(0, 0.05, 0.1, 0.15, 0.2),
rho = c(0.9, 0.95, 0.99)
)
# GBM
gbm_params <- list(
col_sample_rate = c(0.4, 0.7, 1.0),
col_sample_rate_per_tree = c(0.4, 0.7, 1.0),
learn_rate = 0.1,
max_depth = c(3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17),
min_rows = c(1, 5, 10, 15, 30, 100),
min_split_improvement = c(1e-4, 1e-5),
ntrees = 10000, # early stopping
sample_rate = c(0.5, 0.6, 0.7, 0.8, 0.9, 1.0)
)
# XGBoost
xgboost_params <- list(
booster = c("gbtree", "dart"),
col_sample_rate = c(0.6, 0.8, 1.0),
col_sample_rate_per_tree = c(0.7, 0.8, 0.9, 1.0),
max_depth = c(5, 10, 15, 20),
min_rows = c(0.01, 0.1, 1.0, 3.0, 5.0, 10.0, 15.0, 20.0),
ntrees = 10000, # early stopping
reg_alpha = c(0.001, 0.01, 0.1, 1, 10, 100),
reg_lambda = c(0.001, 0.01, 0.1, 0.5, 1),
sample_rate = c(0.6, 0.8, 1.0)
)balanced_accuracy <- function(y_pred, y_true) {
return(mean(c(Sensitivity(y_pred, y_true), Specificity(y_pred, y_true)), na.rm = TRUE))
}
sem <- function(x, na.rm = TRUE) sd(x, na.rm) / sqrt(length(na.omit(x)))
statistical_testing <- function(data, metric_vec, decreasing_vec, p_value_threshold = 0.05, num_entries = 10,
output_path = NULL) {
for (i in 1:length(metric_vec)) {
metric <- metric_vec[i]
ranked_models <- rownames(data[order(data[, paste(metric, "mean")], decreasing = decreasing_vec[i]), ])
pval <- c()
for (j in 2:length(ranked_models)) {
if (sum(is.na(data[ranked_models[j], paste(metric, "CV", 1:10)]))) {
pval <- c(pval, NA)
} else {
pval <- c(pval, wilcox.test(as.numeric(data[ranked_models[1], paste(metric, "CV", 1:10)]),
as.numeric(data[ranked_models[j], paste(metric, "CV", 1:10)]),
exact = FALSE
)$p.value)
}
}
adj_pval <- c(NA, p.adjust(pval, method = "BH", n = length(pval)))
df <- data.frame(adj_pval)
rownames(df) <- ranked_models
colnames(df) <- paste(metric, "adj. <i>p</i> value")
data <- merge(data, df, by = "row.names", all = TRUE)
rownames(data) <- data$Row.names
data$Row.names <- NULL
}
for (i in 1:length(metric_vec)) {
if (decreasing_vec[i]) {
data[paste(metric_vec[i], "rank")] <- rank(-data[, paste(metric_vec[i], "mean")], ties.method = "average")
} else {
data[paste(metric_vec[i], "rank")] <- rank(data[, paste(metric_vec[i], "mean")], ties.method = "average")
}
}
data["Rank sum"] <- rowSums(data[(ncol(data) - length(metric_vec) + 1):ncol(data)])
data <- data[order(data$`Rank sum`), ]
competitive <- rowSums(data[paste(metric_vec, "adj. <i>p</i> value")] < p_value_threshold) == 0
competitive[is.na(competitive)] <- TRUE
data <- data[competitive, ]
if (!is.null(output_path)) {
data[c(
paste(metric_vec, "adj. <i>p</i> value"), paste(metric_vec, "rank"), "Rank sum",
melt(sapply(metric_vec, function(x) paste(x, c("mean", "s.e.m."))))$value
)] %>%
round(digits = 4) %>%
write.csv(., file = output_path)
}
data[c(paste(metric_vec, "adj. <i>p</i> value"), paste(metric_vec, "rank"), "Rank sum")] %>%
round(digits = 4) %>%
datatable(options = list(pageLength = num_entries), escape = FALSE)
}5.2 Melanin binding (regression)
load(file = "./rdata/var_reduct_mb_train_test_splits.RData")
set.seed(32)
# Shuffle the response variable of cross-validation training sets
for (i in 1:10) {
response <- outer_splits[[i]]$train$log_intensity
response <- sample(response)
outer_splits[[i]]$train$log_intensity <- response
}
# Shuffle the response variable of the whole data set
response <- mb_data$log_intensity
response <- sample(response)
mb_data$log_intensity <- response
save(mb_data, outer_splits,
file = "./rdata/var_reduct_mb_train_test_splits_y_shuffled.RData"
)5.2.1 Model training
train_mb_models <- function(train_set, exp_dir, prefix, nfolds = 10, grid_seed = 1) {
tmp <- as.h2o(train_set, destination_frame = prefix)
y <- "log_intensity"
x <- setdiff(names(tmp), y)
res <- as.data.frame(tmp$log_intensity)
# -------------------
# base model training
# -------------------
cat("Deep learning grid 1\n")
deeplearning_1 <- h2o.grid(
algorithm = "deeplearning", x = x, y = y, training_frame = tmp, seed = 1, nfolds = nfolds,
keep_cross_validation_predictions = TRUE, hyper_params = deeplearning_params_1,
stopping_rounds = 3, keep_cross_validation_models = FALSE,
search_criteria = list(
strategy = "RandomDiscrete",
max_models = 100, seed = grid_seed
),
fold_assignment = "Modulo", parallelism = 0
)
for (model_id in deeplearning_1@model_ids) {
tmp_path <- h2o.saveModel(h2o.getModel(model_id), path = exp_dir, force = TRUE)
}
cat("Deep learning grid 2\n")
deeplearning_2 <- h2o.grid(
algorithm = "deeplearning", x = x, y = y, training_frame = tmp, seed = 1, nfolds = nfolds,
keep_cross_validation_predictions = TRUE, hyper_params = deeplearning_params_2,
stopping_rounds = 3, keep_cross_validation_models = FALSE,
search_criteria = list(
strategy = "RandomDiscrete",
max_models = 100, seed = grid_seed
),
fold_assignment = "Modulo", parallelism = 0
)
for (model_id in deeplearning_2@model_ids) {
tmp_path <- h2o.saveModel(h2o.getModel(model_id), path = exp_dir, force = TRUE)
}
cat("Deep learning grid 3\n")
deeplearning_3 <- h2o.grid(
algorithm = "deeplearning", x = x, y = y, training_frame = tmp, seed = 1, nfolds = nfolds,
keep_cross_validation_predictions = TRUE, hyper_params = deeplearning_params_3,
stopping_rounds = 3, keep_cross_validation_models = FALSE,
search_criteria = list(
strategy = "RandomDiscrete",
max_models = 100, seed = grid_seed
),
fold_assignment = "Modulo", parallelism = 0
)
for (model_id in deeplearning_3@model_ids) {
tmp_path <- h2o.saveModel(h2o.getModel(model_id), path = exp_dir, force = TRUE)
}
cat("GBM grid\n")
gbm <- h2o.grid(
algorithm = "gbm", x = x, y = y, training_frame = tmp, seed = 1, nfolds = nfolds,
keep_cross_validation_predictions = TRUE, hyper_params = gbm_params, stopping_rounds = 3,
search_criteria = list(strategy = "RandomDiscrete", max_models = 300, seed = grid_seed),
keep_cross_validation_models = FALSE, fold_assignment = "Modulo", parallelism = 0
)
for (model_id in gbm@model_ids) {
tmp_path <- h2o.saveModel(h2o.getModel(model_id), path = exp_dir, force = TRUE)
}
cat("GBM 5 default models\n")
gbm_1 <- h2o.gbm(
model_id = "GBM_1", x = x, y = y, training_frame = tmp, seed = 1, nfolds = nfolds,
keep_cross_validation_predictions = TRUE, stopping_rounds = 3, score_tree_interval = 5,
col_sample_rate = 0.8, col_sample_rate_per_tree = 0.8, learn_rate = 0.1,
max_depth = 6, min_rows = 1, min_split_improvement = 1e-5, ntrees = 10000, sample_rate = 0.8,
keep_cross_validation_models = FALSE, fold_assignment = "Modulo"
)
gbm_2 <- h2o.gbm(
model_id = "GBM_2", x = x, y = y, training_frame = tmp, seed = 1, nfolds = nfolds,
keep_cross_validation_predictions = TRUE, stopping_rounds = 3, score_tree_interval = 5,
col_sample_rate = 0.8, col_sample_rate_per_tree = 0.8, learn_rate = 0.1,
max_depth = 7, min_rows = 10, min_split_improvement = 1e-5, ntrees = 10000, sample_rate = 0.8,
keep_cross_validation_models = FALSE, fold_assignment = "Modulo"
)
gbm_3 <- h2o.gbm(
model_id = "GBM_3", x = x, y = y, training_frame = tmp, seed = 1, nfolds = nfolds,
keep_cross_validation_predictions = TRUE, stopping_rounds = 3, score_tree_interval = 5,
col_sample_rate = 0.8, col_sample_rate_per_tree = 0.8, learn_rate = 0.1,
max_depth = 8, min_rows = 10, min_split_improvement = 1e-5, ntrees = 10000, sample_rate = 0.8,
keep_cross_validation_models = FALSE, fold_assignment = "Modulo"
)
gbm_4 <- h2o.gbm(
model_id = "GBM_4", x = x, y = y, training_frame = tmp, seed = 1, nfolds = nfolds,
keep_cross_validation_predictions = TRUE, stopping_rounds = 3, score_tree_interval = 5,
col_sample_rate = 0.8, col_sample_rate_per_tree = 0.8, learn_rate = 0.1,
max_depth = 10, min_rows = 10, min_split_improvement = 1e-5, ntrees = 10000, sample_rate = 0.8,
keep_cross_validation_models = FALSE, fold_assignment = "Modulo"
)
gbm_5 <- h2o.gbm(
model_id = "GBM_5", x = x, y = y, training_frame = tmp, seed = 1, nfolds = nfolds,
keep_cross_validation_predictions = TRUE, stopping_rounds = 3, score_tree_interval = 5,
col_sample_rate = 0.8, col_sample_rate_per_tree = 0.8, learn_rate = 0.1,
max_depth = 15, min_rows = 100, min_split_improvement = 1e-5, ntrees = 10000, sample_rate = 0.8,
keep_cross_validation_models = FALSE, fold_assignment = "Modulo"
)
for (model_id in paste0("GBM_", 1:5)) {
tmp_path <- h2o.saveModel(h2o.getModel(model_id), path = exp_dir, force = TRUE)
}
cat("XGBoost grid\n")
xgboost <- h2o.grid(
algorithm = "xgboost", x = x, y = y, training_frame = tmp, seed = 1, nfolds = nfolds,
keep_cross_validation_predictions = TRUE, hyper_params = xgboost_params, stopping_rounds = 3,
search_criteria = list(strategy = "RandomDiscrete", max_models = 300, seed = grid_seed),
keep_cross_validation_models = FALSE, fold_assignment = "Modulo", parallelism = 0
)
for (model_id in xgboost@model_ids) {
tmp_path <- h2o.saveModel(h2o.getModel(model_id), path = exp_dir, force = TRUE)
}
cat("XGBoost 3 default models\n")
xgboost_1 <- h2o.xgboost(
model_id = "XGBoost_1", x = x, y = y, training_frame = tmp, seed = 1, nfolds = nfolds,
keep_cross_validation_predictions = TRUE, stopping_rounds = 3, score_tree_interval = 5,
booster = "gbtree", col_sample_rate = 0.8, col_sample_rate_per_tree = 0.8,
max_depth = 10, min_rows = 5, ntrees = 10000, reg_alpha = 0, reg_lambda = 1,
sample_rate = 0.6, keep_cross_validation_models = FALSE, fold_assignment = "Modulo"
)
xgboost_2 <- h2o.xgboost(
model_id = "XGBoost_2", x = x, y = y, training_frame = tmp, seed = 1, nfolds = nfolds,
keep_cross_validation_predictions = TRUE, stopping_rounds = 3, score_tree_interval = 5,
booster = "gbtree", col_sample_rate = 0.8, col_sample_rate_per_tree = 0.8,
max_depth = 20, min_rows = 10, ntrees = 10000, reg_alpha = 0, reg_lambda = 1,
sample_rate = 0.6, keep_cross_validation_models = FALSE, fold_assignment = "Modulo"
)
xgboost_3 <- h2o.xgboost(
model_id = "XGBoost_3", x = x, y = y, training_frame = tmp, seed = 1, nfolds = nfolds,
keep_cross_validation_predictions = TRUE, stopping_rounds = 3, score_tree_interval = 5,
booster = "gbtree", col_sample_rate = 0.8, col_sample_rate_per_tree = 0.8,
max_depth = 5, min_rows = 3, ntrees = 10000, reg_alpha = 0, reg_lambda = 1,
sample_rate = 0.8, keep_cross_validation_models = FALSE, fold_assignment = "Modulo"
)
for (model_id in paste0("XGBoost_", 1:3)) {
tmp_path <- h2o.saveModel(h2o.getModel(model_id), path = exp_dir, force = TRUE)
}
cat("GLM\n")
glm <- h2o.glm(
model_id = "GLM", x = x, y = y, training_frame = tmp, seed = 1, nfolds = nfolds,
keep_cross_validation_predictions = TRUE, alpha = c(0.0, 0.2, 0.4, 0.6, 0.8, 1.0),
keep_cross_validation_models = FALSE, fold_assignment = "Modulo"
)
tmp_path <- h2o.saveModel(h2o.getModel("GLM"), path = exp_dir, force = TRUE)
cat("DRF\n")
drf <- h2o.randomForest(
model_id = "DRF", x = x, y = y, training_frame = tmp, seed = 1, nfolds = nfolds,
keep_cross_validation_predictions = TRUE, ntrees = 10000,
score_tree_interval = 5, stopping_rounds = 3,
keep_cross_validation_models = FALSE, fold_assignment = "Modulo"
)
tmp_path <- h2o.saveModel(h2o.getModel("DRF"), path = exp_dir, force = TRUE)
cat("XRT\n")
xrt <- h2o.randomForest(
model_id = "XRT", x = x, y = y, training_frame = tmp, seed = 1, nfolds = nfolds,
keep_cross_validation_predictions = TRUE, ntrees = 10000, histogram_type = "Random",
score_tree_interval = 5, stopping_rounds = 3,
keep_cross_validation_models = FALSE, fold_assignment = "Modulo"
)
tmp_path <- h2o.saveModel(h2o.getModel("XRT"), path = exp_dir, force = TRUE)
# -----------------------
# get holdout predictions
# -----------------------
base_models <- as.list(c(
unlist(deeplearning_1@model_ids),
unlist(deeplearning_2@model_ids),
unlist(deeplearning_3@model_ids),
unlist(gbm@model_ids), paste0("GBM_", 1:5),
unlist(xgboost@model_ids), paste0("XGBoost_", 1:3),
"GLM",
"DRF",
"XRT"
))
for (model_id in base_models) {
res <- cbind(res, as.data.frame(h2o.getFrame(paste0("cv_holdout_prediction_", model_id)),
col.names = model_id
))
}
# ----------------------
# super learner training
# ----------------------
sl_iter <- 0
cat(paste0("Super learner iteration ", sl_iter, " (", length(base_models), " models)\n"))
sl <- h2o.stackedEnsemble(
x = x, y = y, model_id = paste0("superlearner_iter_", sl_iter),
training_frame = tmp, seed = 1,
base_models = base_models,
metalearner_algorithm = "glm",
metalearner_nfolds = nfolds,
keep_levelone_frame = TRUE,
metalearner_params = list(standardize = TRUE, keep_cross_validation_predictions = TRUE)
)
tmp_path <- h2o.saveModel(h2o.getModel(paste0("superlearner_iter_", sl_iter)), path = exp_dir, force = TRUE)
model_id <- paste0("metalearner_glm_superlearner_iter_", sl_iter)
tmp_path <- h2o.saveModel(h2o.getModel(model_id), path = exp_dir, force = TRUE)
res <- cbind(res, as.data.frame(h2o.getFrame(paste0("cv_holdout_prediction_", model_id)),
col.names = paste0(model_id, "_", length(base_models), "_models")
))
# ----------------------------------
# super learner base model reduction
# ----------------------------------
while (TRUE) {
meta <- h2o.getModel(paste0("metalearner_glm_superlearner_iter_", sl_iter, "_cv_1"))
names <- meta@model$coefficients_table[, "names"]
coeffs <- (meta@model$coefficients_table[, "standardized_coefficients"] > 0)
for (j in 2:nfolds) {
meta <- h2o.getModel(paste0("metalearner_glm_superlearner_iter_", sl_iter, "_cv_", j))
names <- meta@model$coefficients_table[, "names"]
coeffs <- coeffs + (meta@model$coefficients_table[, "standardized_coefficients"] > 0)
}
base_models_ <- as.list(names[coeffs >= ceiling(nfolds / 2) & names != "Intercept"])
if (length(base_models_) == 0) {
cat("No base models passing the threshold\n\n")
break
}
if (sum(base_models %in% base_models_) == length(base_models)) {
cat("No further reduction of base models\n\n")
break
}
sl_iter <- sl_iter + 1
base_models <- base_models_
cat(paste0("Super learner iteration ", sl_iter, " (", length(base_models), " models)\n"))
sl <- h2o.stackedEnsemble(
x = x, y = y, model_id = paste0("superlearner_iter_", sl_iter),
training_frame = tmp, seed = 1,
base_models = base_models,
metalearner_algorithm = "glm",
metalearner_nfolds = nfolds,
keep_levelone_frame = TRUE,
metalearner_params = list(standardize = TRUE, keep_cross_validation_predictions = TRUE)
)
tmp_path <- h2o.saveModel(h2o.getModel(paste0("superlearner_iter_", sl_iter)), path = exp_dir, force = TRUE)
model_id <- paste0("metalearner_glm_superlearner_iter_", sl_iter)
tmp_path <- h2o.saveModel(h2o.getModel(model_id), path = exp_dir, force = TRUE)
res <- cbind(res, as.data.frame(h2o.getFrame(paste0("cv_holdout_prediction_", model_id)),
col.names = paste0(model_id, "_", length(base_models), "_models")
))
}
# -----------------------------------------
# super learner for homogeneous base models
# -----------------------------------------
# DeepLearning
base_models <- as.list(c(
unlist(deeplearning_1@model_ids),
unlist(deeplearning_2@model_ids),
unlist(deeplearning_3@model_ids)
))
cat(paste0("Super learner deep learning (", length(base_models), " models)\n"))
sl <- h2o.stackedEnsemble(
x = x, y = y, model_id = "superlearner_deeplearning",
training_frame = tmp, seed = 1,
base_models = base_models,
metalearner_algorithm = "glm",
metalearner_nfolds = nfolds,
keep_levelone_frame = TRUE,
metalearner_params = list(standardize = TRUE, keep_cross_validation_predictions = TRUE)
)
tmp_path <- h2o.saveModel(h2o.getModel("superlearner_deeplearning"), path = exp_dir, force = TRUE)
model_id <- "metalearner_glm_superlearner_deeplearning"
tmp_path <- h2o.saveModel(h2o.getModel(model_id), path = exp_dir, force = TRUE)
res <- cbind(res, as.data.frame(h2o.getFrame(paste0("cv_holdout_prediction_", model_id)),
col.names = paste0(model_id, "_", length(base_models), "_models")
))
# GBM
base_models <- as.list(c(unlist(gbm@model_ids), paste0("GBM_", 1:5)))
cat(paste0("Super learner GBM (", length(base_models), " models)\n"))
sl <- h2o.stackedEnsemble(
x = x, y = y, model_id = "superlearner_gbm",
training_frame = tmp, seed = 1,
base_models = base_models,
metalearner_algorithm = "glm",
metalearner_nfolds = nfolds,
keep_levelone_frame = TRUE,
metalearner_params = list(standardize = TRUE, keep_cross_validation_predictions = TRUE)
)
tmp_path <- h2o.saveModel(h2o.getModel("superlearner_gbm"), path = exp_dir, force = TRUE)
model_id <- "metalearner_glm_superlearner_gbm"
tmp_path <- h2o.saveModel(h2o.getModel(model_id), path = exp_dir, force = TRUE)
res <- cbind(res, as.data.frame(h2o.getFrame(paste0("cv_holdout_prediction_", model_id)),
col.names = paste0(model_id, "_", length(base_models), "_models")
))
# XGBoost
base_models <- as.list(c(unlist(xgboost@model_ids), paste0("XGBoost_", 1:3)))
cat(paste0("Super learner XGBoost (", length(base_models), " models)\n"))
sl <- h2o.stackedEnsemble(
x = x, y = y, model_id = "superlearner_xgboost",
training_frame = tmp, seed = 1,
base_models = base_models,
metalearner_algorithm = "glm",
metalearner_nfolds = nfolds,
keep_levelone_frame = TRUE,
metalearner_params = list(standardize = TRUE, keep_cross_validation_predictions = TRUE)
)
tmp_path <- h2o.saveModel(h2o.getModel("superlearner_xgboost"), path = exp_dir, force = TRUE)
model_id <- "metalearner_glm_superlearner_xgboost"
tmp_path <- h2o.saveModel(h2o.getModel(model_id), path = exp_dir, force = TRUE)
res <- cbind(res, as.data.frame(h2o.getFrame(paste0("cv_holdout_prediction_", model_id)),
col.names = paste0(model_id, "_", length(base_models), "_models")
))
write.csv(res, file = paste0(exp_dir, "/cv_holdout_predictions.csv"), row.names = FALSE)
cat("\n\n")
h2o.removeAll()
}5.2.2 Inner cross-validation
load(file = "./rdata/var_reduct_mb_train_test_splits_y_shuffled.RData")
for (i in 1:10) {
cat(paste0("Outer training set ", i, "\n"))
prefix <- paste0("outer_", i)
exp_dir <- paste0("/Users/renee/Downloads/melanin_binding_adversarial/", prefix)
dir.create(exp_dir)
train_mb_models(train_set = outer_splits[[i]][["train"]], exp_dir = exp_dir, prefix = prefix)
}# Keep track of grid models
dl_grid <- list()
gbm_grid <- list()
xgboost_grid <- list()
dl_grid_params <- list()
gbm_grid_params <- list()
xgboost_grid_params <- list()
for (i in 1:10) {
cat(paste0("Outer training set ", i, "\n"))
prefix <- paste0("outer_", i)
dir <- paste0("/Users/renee/Downloads/melanin_binding_adversarial/", prefix)
files <- list.files(dir)
# Deep learning
dl <- files[str_detect(files, "DeepLearning_model")]
for (m in dl) {
model <- h2o.loadModel(paste0(dir, "/", m))
hs <- sha1(paste(c(
model@allparameters$epsilon,
model@allparameters$hidden,
model@allparameters$hidden_dropout_ratios,
model@allparameters$input_dropout_ratio,
model@allparameters$rho
), collapse = " "))
if (hs %in% names(dl_grid)) {
dl_grid[[hs]] <- c(dl_grid[[hs]], m)
} else {
dl_grid[[hs]] <- c(m)
dl_grid_params <- list.append(
dl_grid_params,
c(
"epsilon" = model@allparameters$epsilon,
"hidden" = paste0("[", paste(model@allparameters$hidden, collapse = ","), "]"),
"hidden_dropout_ratios" = paste0(
"[",
paste(model@allparameters$hidden_dropout_ratios,
collapse = ","
), "]"
),
"input_dropout_ratio" = model@allparameters$input_dropout_ratio,
"rho" = model@allparameters$rho
)
)
}
}
h2o.removeAll()
# GBM
gbm <- files[str_detect(files, "GBM_model")]
for (m in gbm) {
model <- h2o.loadModel(paste0(dir, "/", m))
hs <- sha1(paste(c(
model@allparameters$col_sample_rate,
model@allparameters$col_sample_rate_per_tree,
model@allparameters$max_depth,
model@allparameters$min_rows,
model@allparameters$min_split_improvement,
model@allparameters$sample_rate
), collapse = " "))
if (hs %in% names(gbm_grid)) {
gbm_grid[[hs]] <- c(gbm_grid[[hs]], m)
} else {
gbm_grid[[hs]] <- c(m)
gbm_grid_params <- list.append(
gbm_grid_params,
c(
"col_sample_rate" = model@allparameters$col_sample_rate,
"col_sample_rate_per_tree" = model@allparameters$col_sample_rate_per_tree,
"max_depth" = model@allparameters$max_depth,
"min_rows" = model@allparameters$min_rows,
"min_split_improvement" = model@allparameters$min_split_improvement,
"sample_rate" = model@allparameters$sample_rate
)
)
}
}
h2o.removeAll()
# XGBoost
xgboost <- files[str_detect(files, "XGBoost_model")]
for (m in xgboost) {
model <- h2o.loadModel(paste0(dir, "/", m))
hs <- sha1(paste(c(
model@allparameters$booster,
model@allparameters$col_sample_rate,
model@allparameters$col_sample_rate_per_tree,
model@allparameters$max_depth,
model@allparameters$min_rows,
model@allparameters$reg_alpha,
model@allparameters$reg_lambda,
model@allparameters$sample_rate
), collapse = " "))
if (hs %in% names(xgboost_grid)) {
xgboost_grid[[hs]] <- c(xgboost_grid[[hs]], m)
} else {
xgboost_grid[[hs]] <- c(m)
xgboost_grid_params <- list.append(
xgboost_grid_params,
c(
"booster" = model@allparameters$booster,
"col_sample_rate" = model@allparameters$col_sample_rate,
"col_sample_rate_per_tree" = model@allparameters$col_sample_rate_per_tree,
"max_depth" = model@allparameters$max_depth,
"min_rows" = model@allparameters$min_rows,
"reg_alpha" = model@allparameters$reg_alpha,
"reg_lambda" = model@allparameters$reg_lambda,
"sample_rate" = model@allparameters$sample_rate
)
)
}
}
h2o.removeAll()
}
dl_grid_params <- as.data.frame(t(data.frame(dl_grid_params)))
rownames(dl_grid_params) <- paste("Neural network grid model", 1:nrow(dl_grid_params))
gbm_grid_params <- as.data.frame(t(data.frame(gbm_grid_params)))
rownames(gbm_grid_params) <- paste("GBM grid model", 1:nrow(gbm_grid_params))
xgboost_grid_params <- as.data.frame(t(data.frame(xgboost_grid_params)))
rownames(xgboost_grid_params) <- paste("XGBoost grid model", 1:nrow(xgboost_grid_params))
write.csv(dl_grid_params, "./other_data/melanin_binding_adversarial/neural_network_grid_params.csv")
write.csv(gbm_grid_params, "./other_data/melanin_binding_adversarial/gbm_grid_params.csv")
write.csv(xgboost_grid_params, "./other_data/melanin_binding_adversarial/xgboost_grid_params.csv")
save(dl_grid, gbm_grid, xgboost_grid, file = "./rdata/model_training_grid_models_mb_ad.RData")5.2.3 Model evaluation
model_evaluation <- function(holdout_pred, fold_asign, grid_name, grid_meta = NULL) {
load(file = "./rdata/var_reduct_mb_train_test_splits_y_shuffled.RData")
res_ <- list()
model_num <- c()
if (startsWith(grid_name, "Super learner")) {
if (grid_name == "Super learner all models") {
m <- colnames(holdout_pred)[startsWith(colnames(holdout_pred), "metalearner_glm_superlearner_iter_0")]
} else if (grid_name == "Super learner final") {
sl_models <- colnames(holdout_pred)[startsWith(colnames(holdout_pred), "metalearner_glm_superlearner_iter")]
m <- sl_models[length(sl_models)]
} else if (grid_name == "Super learner neural network") {
m <- colnames(holdout_pred)[startsWith(colnames(holdout_pred), "metalearner_glm_superlearner_deeplearning")]
} else if (grid_name == "Super learner GBM") {
m <- colnames(holdout_pred)[startsWith(colnames(holdout_pred), "metalearner_glm_superlearner_gbm")]
} else if (grid_name == "Super learner XGBoost") {
m <- colnames(holdout_pred)[startsWith(colnames(holdout_pred), "metalearner_glm_superlearner_xgboost")]
}
mae <- c()
rmse <- c()
R2 <- c()
for (k in 1:10) {
y_true <- holdout_pred[fold_assign == k, "log_intensity"]
y_pred <- holdout_pred[fold_assign == k, m]
mae <- c(mae, MAE(y_pred = y_pred, y_true = y_true))
rmse <- c(rmse, RMSE(y_pred = y_pred, y_true = y_true))
R2 <- c(R2, R2_Score(y_pred = y_pred, y_true = y_true))
}
# Re-scale to 0-100
mae <- rescale(mae, to = c(0, 100), from = range(mb_data$log_intensity))
rmse <- rescale(rmse, to = c(0, 100), from = range(mb_data$log_intensity))
res_ <- list.append(res_, c(
mean(mae, na.rm = TRUE), sem(mae),
mean(rmse, na.rm = TRUE), sem(rmse),
mean(R2, na.rm = TRUE), sem(R2),
mae, rmse, R2
))
} else {
for (j in 1:length(grid_meta)) {
g <- grid_meta[[j]]
m <- intersect(colnames(holdout_pred), g)
if (length(m) == 1) {
model_num <- c(model_num, j)
mae <- c()
rmse <- c()
R2 <- c()
for (k in 1:10) {
y_true <- holdout_pred[fold_assign == k, "log_intensity"]
y_pred <- holdout_pred[fold_assign == k, m]
mae <- c(mae, MAE(y_pred = y_pred, y_true = y_true))
rmse <- c(rmse, RMSE(y_pred = y_pred, y_true = y_true))
R2 <- c(R2, R2_Score(y_pred = y_pred, y_true = y_true))
}
# Re-scale to 0-100
mae <- rescale(mae, to = c(0, 100), from = range(mb_data$log_intensity))
rmse <- rescale(rmse, to = c(0, 100), from = range(mb_data$log_intensity))
res_ <- list.append(res_, c(
mean(mae, na.rm = TRUE), sem(mae),
mean(rmse, na.rm = TRUE), sem(rmse),
mean(R2, na.rm = TRUE), sem(R2),
mae, rmse, R2
))
}
}
}
res <- as.data.frame(t(data.frame(res_)))
colnames(res) <- c(
"Norm. MAE mean", "Norm. MAE s.e.m.", "Norm. RMSE mean", "Norm. RMSE s.e.m.",
"R^2 mean", "R^2 s.e.m.",
paste("Norm. MAE CV", 1:10), paste("Norm. RMSE CV", 1:10), paste("R^2 CV", 1:10)
)
if (nrow(res) == 1) {
rownames(res) <- grid_name
} else {
rownames(res) <- paste(grid_name, model_num)
}
return(res)
}5.2.4 Inner loop model selection
load(file = "./rdata/model_training_grid_models_mb_ad.RData")
for (i in 1:10) {
holdout_pred <- read.csv(paste0("./other_data/melanin_binding_adversarial/outer_", i, "/cv_holdout_predictions.csv"))
fold_assign <- rep(1:10, ceiling(nrow(holdout_pred) / 10))[1:nrow(holdout_pred)]
data_ <- list()
# Deep learning grid models
data_ <- list.append(data_, model_evaluation(holdout_pred, fold_assign,
grid_name = "Neural network grid model",
grid_meta = dl_grid
))
# GBM grid models
data_ <- list.append(data_, model_evaluation(holdout_pred, fold_assign,
grid_name = "GBM grid model",
grid_meta = gbm_grid
))
# GBM default models
data_ <- list.append(data_, model_evaluation(holdout_pred, fold_assign,
grid_name = "GBM model",
grid_meta = as.list(paste0("GBM_", 1:5))
))
# XGBoost grid models
data_ <- list.append(data_, model_evaluation(holdout_pred, fold_assign,
grid_name = "XGBoost grid model",
grid_meta = xgboost_grid
))
# XGBoost default models
data_ <- list.append(data_, model_evaluation(holdout_pred, fold_assign,
grid_name = "XGBoost model",
grid_meta = as.list(paste0("XGBoost_", 1:3))
))
# GLM
data_ <- list.append(data_, model_evaluation(holdout_pred, fold_assign, grid_name = "GLM model", grid_meta = list("GLM")))
# DRF
data_ <- list.append(data_, model_evaluation(holdout_pred, fold_assign, grid_name = "DRF model", grid_meta = list("DRF")))
# XRT
data_ <- list.append(data_, model_evaluation(holdout_pred, fold_assign, grid_name = "XRT model", grid_meta = list("XRT")))
# Super learner all
data_ <- list.append(data_, model_evaluation(holdout_pred, fold_assign, grid_name = "Super learner all models"))
# Super learner final
data_ <- list.append(data_, model_evaluation(holdout_pred, fold_assign, grid_name = "Super learner final"))
# Super learner deep learning
data_ <- list.append(data_, model_evaluation(holdout_pred, fold_assign, grid_name = "Super learner neural network"))
# Super learner GBM
data_ <- list.append(data_, model_evaluation(holdout_pred, fold_assign, grid_name = "Super learner GBM"))
# Super learner XGBoost
data_ <- list.append(data_, model_evaluation(holdout_pred, fold_assign, grid_name = "Super learner XGBoost"))
data <- do.call(rbind, data_)
write.csv(data, paste0("./other_data/melanin_binding_adversarial/outer_", i, "/cv_res.csv"))
}5.2.5 Final evaluation
load(file = "./rdata/var_reduct_mb_train_test_splits_y_shuffled.RData")
load(file = "./rdata/model_training_grid_models_mb_ad.RData")
dir <- "/Users/renee/Downloads/melanin_binding_adversarial"
selected_models <- c(
"Neural network grid model 113", "Neural network grid model 187",
"Neural network grid model 145", "Neural network grid model 215",
"Super learner final", "Neural network grid model 188",
"Neural network grid model 176", "Neural network grid model 201",
"Neural network grid model 139", "Neural network grid model 160"
)
mae <- c()
rmse <- c()
R2 <- c()
for (i in 1:10) {
holdout_pred <- read.csv(paste0("./other_data/melanin_binding_adversarial/outer_", i, "/cv_holdout_predictions.csv"))
if (startsWith(selected_models[i], "Neural network grid model")) {
grid_num <- as.integer(str_extract(selected_models[i], "[0-9]+"))
m <- intersect(colnames(holdout_pred), dl_grid[[grid_num]])
model <- h2o.loadModel(paste0(dir, "/outer_", i, "/", m))
} else if (startsWith(selected_models[i], "GBM grid model")) {
grid_num <- as.integer(str_extract(selected_models[i], "[0-9]+"))
m <- intersect(colnames(holdout_pred), gbm_grid[[grid_num]])
model <- h2o.loadModel(paste0(dir, "/outer_", i, "/", m))
} else if (startsWith(selected_models[i], "GBM model")) {
grid_num <- as.integer(str_extract(selected_models[i], "[0-9]+"))
model <- h2o.loadModel(paste0(dir, "/outer_", i, "/GBM_", grid_num))
} else if (startsWith(selected_models[i], "XGBoost grid model")) {
grid_num <- as.integer(str_extract(selected_models[i], "[0-9]+"))
m <- intersect(colnames(holdout_pred), xgboost_grid[[grid_num]])
model <- h2o.loadModel(paste0(dir, "/outer_", i, "/", m))
} else if (startsWith(selected_models[i], "XGBoost model")) {
grid_num <- as.integer(str_extract(selected_models[i], "[0-9]+"))
model <- h2o.loadModel(paste0(dir, "/outer_", i, "/XGBoost_", grid_num))
} else if (selected_models[i] == "Super learner all models") {
model <- h2o.loadModel(paste0(dir, "/outer_", i, "/superlearner_iter_0"))
} else if (selected_models[i] == "Super learner final") {
files <- list.files(paste0(dir, "/outer_", i))
files <- files[startsWith(files, "superlearner_iter")]
model <- h2o.loadModel(paste0(dir, "/outer_", i, "/", files[length(files)]))
} else if (selected_models[i] == "Super learner neural network") {
model <- h2o.loadModel(paste0(dir, "/outer_", i, "/superlearner_deeplearning"))
} else if (selected_models[i] == "Super learner GBM") {
model <- h2o.loadModel(paste0(dir, "/outer_", i, "/superlearner_gbm"))
} else if (selected_models[i] == "Super learner XGBoost") {
model <- h2o.loadModel(paste0(dir, "/outer_", i, "/superlearner_xgboost"))
} else {
model <- h2o.loadModel(paste0(dir, "/outer_", i, "/", unlist(strsplit(selected_models[i], " "))[1]))
}
tmp <- as.h2o(subset(outer_splits[[i]][["test"]], select = -log_intensity))
y_true <- outer_splits[[i]][["test"]]$log_intensity
y_pred <- as.data.frame(as.data.frame(h2o.predict(model, tmp)))[, 1]
mae <- c(mae, MAE(y_pred = y_pred, y_true = y_true))
rmse <- c(rmse, RMSE(y_pred = y_pred, y_true = y_true))
R2 <- c(R2, R2_Score(y_pred = y_pred, y_true = y_true))
}
# Re-scale to 0-100
mae <- rescale(mae, to = c(0, 100), from = range(mb_data$log_intensity))
rmse <- rescale(rmse, to = c(0, 100), from = range(mb_data$log_intensity))
h2o.removeAll()
save(mae, rmse, R2, file = "./rdata/final_evaluation_mb_ad.RData")load(file = "./rdata/final_evaluation_mb_ad.RData")
data.frame(
Metric = c("Norm. MAE", "Norm. RMSE", "R<sup>2</sup>"),
`Mean ± s.e.m.` = c(
paste0(round(mean(mae), 3), " ± ", round(sem(mae), 3)),
paste0(round(mean(rmse), 3), " ± ", round(sem(rmse), 3)),
paste0(round(mean(R2), 3), " ± ", round(sem(R2), 3))
),
check.names = FALSE
) %>%
datatable(escape = FALSE, rownames = FALSE)5.3 Cell-penetration (classification)
load(file = "./rdata/var_reduct_cpp_train_test_splits.RData")
set.seed(32)
# Shuffle the response variable of cross-validation training sets
for (i in 1:10) {
response <- outer_splits[[i]]$train$category
response <- sample(response)
outer_splits[[i]]$train$category <- response
}
# Shuffle the response variable of the whole data set
response <- cpp_data$category
response <- sample(response)
cpp_data$category <- response
save(cpp_data, outer_splits,
file = "./rdata/var_reduct_cpp_train_test_splits_y_shuffled.RData"
)5.3.1 Model training
train_cpp_models <- function(train_set, exp_dir, prefix, nfolds = 10, grid_seed = 1) {
tmp <- as.h2o(train_set, destination_frame = prefix)
tmp["category"] <- as.factor(tmp["category"])
y <- "category"
x <- setdiff(names(tmp), y)
res <- as.data.frame(tmp$category)
samp_factors <- as.vector(mean(table(train_set$category)) / table(train_set$category))
# -------------------
# base model training
# -------------------
cat("Deep learning grid 1\n")
deeplearning_1 <- h2o.grid(
algorithm = "deeplearning", x = x, y = y, training_frame = tmp, seed = 1, nfolds = nfolds,
keep_cross_validation_predictions = TRUE, hyper_params = deeplearning_params_1,
stopping_rounds = 3, balance_classes = TRUE, class_sampling_factors = samp_factors,
search_criteria = list(
strategy = "RandomDiscrete",
max_models = 100, seed = grid_seed
),
keep_cross_validation_models = FALSE, fold_assignment = "Modulo", parallelism = 0
)
for (model_id in deeplearning_1@model_ids) {
tmp_path <- h2o.saveModel(h2o.getModel(model_id), path = exp_dir, force = TRUE)
}
cat("GBM grid\n")
gbm <- h2o.grid(
algorithm = "gbm", x = x, y = y, training_frame = tmp, seed = 1, nfolds = nfolds,
keep_cross_validation_predictions = TRUE, hyper_params = gbm_params, stopping_rounds = 3,
balance_classes = TRUE, class_sampling_factors = samp_factors,
search_criteria = list(strategy = "RandomDiscrete", max_models = 100, seed = grid_seed),
keep_cross_validation_models = FALSE, fold_assignment = "Modulo", parallelism = 0
)
for (model_id in gbm@model_ids) {
tmp_path <- h2o.saveModel(h2o.getModel(model_id), path = exp_dir, force = TRUE)
}
cat("GBM 5 default models\n")
gbm_1 <- h2o.gbm(
model_id = "GBM_1", x = x, y = y, training_frame = tmp, seed = 1, nfolds = nfolds,
keep_cross_validation_predictions = TRUE, stopping_rounds = 3, score_tree_interval = 5,
balance_classes = TRUE, class_sampling_factors = samp_factors,
col_sample_rate = 0.8, col_sample_rate_per_tree = 0.8, learn_rate = 0.1,
max_depth = 6, min_rows = 1, min_split_improvement = 1e-5, ntrees = 10000, sample_rate = 0.8,
keep_cross_validation_models = FALSE, fold_assignment = "Modulo"
)
gbm_2 <- h2o.gbm(
model_id = "GBM_2", x = x, y = y, training_frame = tmp, seed = 1, nfolds = nfolds,
keep_cross_validation_predictions = TRUE, stopping_rounds = 3, score_tree_interval = 5,
balance_classes = TRUE, class_sampling_factors = samp_factors,
col_sample_rate = 0.8, col_sample_rate_per_tree = 0.8, learn_rate = 0.1,
max_depth = 7, min_rows = 10, min_split_improvement = 1e-5, ntrees = 10000, sample_rate = 0.8,
keep_cross_validation_models = FALSE, fold_assignment = "Modulo"
)
gbm_3 <- h2o.gbm(
model_id = "GBM_3", x = x, y = y, training_frame = tmp, seed = 1, nfolds = nfolds,
keep_cross_validation_predictions = TRUE, stopping_rounds = 3, score_tree_interval = 5,
balance_classes = TRUE, class_sampling_factors = samp_factors,
col_sample_rate = 0.8, col_sample_rate_per_tree = 0.8, learn_rate = 0.1,
max_depth = 8, min_rows = 10, min_split_improvement = 1e-5, ntrees = 10000, sample_rate = 0.8,
keep_cross_validation_models = FALSE, fold_assignment = "Modulo"
)
gbm_4 <- h2o.gbm(
model_id = "GBM_4", x = x, y = y, training_frame = tmp, seed = 1, nfolds = nfolds,
keep_cross_validation_predictions = TRUE, stopping_rounds = 3, score_tree_interval = 5,
balance_classes = TRUE, class_sampling_factors = samp_factors,
col_sample_rate = 0.8, col_sample_rate_per_tree = 0.8, learn_rate = 0.1,
max_depth = 10, min_rows = 10, min_split_improvement = 1e-5, ntrees = 10000, sample_rate = 0.8,
keep_cross_validation_models = FALSE, fold_assignment = "Modulo"
)
gbm_5 <- h2o.gbm(
model_id = "GBM_5", x = x, y = y, training_frame = tmp, seed = 1, nfolds = nfolds,
keep_cross_validation_predictions = TRUE, stopping_rounds = 3, score_tree_interval = 5,
balance_classes = TRUE, class_sampling_factors = samp_factors,
col_sample_rate = 0.8, col_sample_rate_per_tree = 0.8, learn_rate = 0.1,
max_depth = 15, min_rows = 100, min_split_improvement = 1e-5, ntrees = 10000, sample_rate = 0.8,
keep_cross_validation_models = FALSE, fold_assignment = "Modulo"
)
for (model_id in paste0("GBM_", 1:5)) {
tmp_path <- h2o.saveModel(h2o.getModel(model_id), path = exp_dir, force = TRUE)
}
cat("XGBoost grid\n")
xgboost <- h2o.grid(
algorithm = "xgboost", x = x, y = y, training_frame = tmp, seed = 1, nfolds = nfolds,
keep_cross_validation_predictions = TRUE, hyper_params = xgboost_params, stopping_rounds = 3,
search_criteria = list(strategy = "RandomDiscrete", max_models = 100, seed = grid_seed),
keep_cross_validation_models = FALSE, fold_assignment = "Modulo", parallelism = 0
)
for (model_id in xgboost@model_ids) {
tmp_path <- h2o.saveModel(h2o.getModel(model_id), path = exp_dir, force = TRUE)
}
cat("XGBoost 3 default models\n")
xgboost_1 <- h2o.xgboost(
model_id = "XGBoost_1", x = x, y = y, training_frame = tmp, seed = 1, nfolds = nfolds,
keep_cross_validation_predictions = TRUE, stopping_rounds = 3, score_tree_interval = 5,
booster = "gbtree", col_sample_rate = 0.8, col_sample_rate_per_tree = 0.8,
max_depth = 10, min_rows = 5, ntrees = 10000, reg_alpha = 0, reg_lambda = 1,
sample_rate = 0.6, keep_cross_validation_models = FALSE, fold_assignment = "Modulo"
)
xgboost_2 <- h2o.xgboost(
model_id = "XGBoost_2", x = x, y = y, training_frame = tmp, seed = 1, nfolds = nfolds,
keep_cross_validation_predictions = TRUE, stopping_rounds = 3, score_tree_interval = 5,
booster = "gbtree", col_sample_rate = 0.8, col_sample_rate_per_tree = 0.8,
max_depth = 20, min_rows = 10, ntrees = 10000, reg_alpha = 0, reg_lambda = 1,
sample_rate = 0.6, keep_cross_validation_models = FALSE, fold_assignment = "Modulo"
)
xgboost_3 <- h2o.xgboost(
model_id = "XGBoost_3", x = x, y = y, training_frame = tmp, seed = 1, nfolds = nfolds,
keep_cross_validation_predictions = TRUE, stopping_rounds = 3, score_tree_interval = 5,
booster = "gbtree", col_sample_rate = 0.8, col_sample_rate_per_tree = 0.8,
max_depth = 5, min_rows = 3, ntrees = 10000, reg_alpha = 0, reg_lambda = 1,
sample_rate = 0.8, keep_cross_validation_models = FALSE, fold_assignment = "Modulo"
)
for (model_id in paste0("XGBoost_", 1:3)) {
tmp_path <- h2o.saveModel(h2o.getModel(model_id), path = exp_dir, force = TRUE)
}
cat("DRF\n")
drf <- h2o.randomForest(
model_id = "DRF", x = x, y = y, training_frame = tmp, seed = 1, nfolds = nfolds,
keep_cross_validation_predictions = TRUE, ntrees = 10000,
score_tree_interval = 5, stopping_rounds = 3,
balance_classes = TRUE, class_sampling_factors = samp_factors,
keep_cross_validation_models = FALSE, fold_assignment = "Modulo"
)
tmp_path <- h2o.saveModel(h2o.getModel("DRF"), path = exp_dir, force = TRUE)
cat("XRT\n")
xrt <- h2o.randomForest(
model_id = "XRT", x = x, y = y, training_frame = tmp, seed = 1, nfolds = nfolds,
keep_cross_validation_predictions = TRUE, ntrees = 10000, histogram_type = "Random",
score_tree_interval = 5, stopping_rounds = 3,
balance_classes = TRUE, class_sampling_factors = samp_factors,
keep_cross_validation_models = FALSE, fold_assignment = "Modulo"
)
tmp_path <- h2o.saveModel(h2o.getModel("XRT"), path = exp_dir, force = TRUE)
# -----------------------
# get holdout predictions
# -----------------------
base_models <- as.list(c(
unlist(deeplearning_1@model_ids),
unlist(gbm@model_ids), paste0("GBM_", 1:5),
unlist(xgboost@model_ids), paste0("XGBoost_", 1:3),
"DRF",
"XRT"
))
for (model_id in base_models) {
res <- cbind(res, as.data.frame(h2o.getFrame(paste0("cv_holdout_prediction_", model_id))["penetrating"],
col.names = model_id
))
}
# ----------------------
# super learner training
# ----------------------
sl_iter <- 0
cat(paste0("Super learner iteration ", sl_iter, " (", length(base_models), " models)\n"))
sl <- h2o.stackedEnsemble(
x = x, y = y, model_id = paste0("superlearner_iter_", sl_iter),
training_frame = tmp, seed = 1,
base_models = base_models,
metalearner_algorithm = "glm",
metalearner_nfolds = nfolds,
keep_levelone_frame = TRUE,
metalearner_params = list(
standardize = TRUE, keep_cross_validation_predictions = TRUE,
balance_classes = TRUE, class_sampling_factors = samp_factors
)
)
tmp_path <- h2o.saveModel(h2o.getModel(paste0("superlearner_iter_", sl_iter)), path = exp_dir, force = TRUE)
model_id <- paste0("metalearner_glm_superlearner_iter_", sl_iter)
tmp_path <- h2o.saveModel(h2o.getModel(model_id), path = exp_dir, force = TRUE)
res <- cbind(res, as.data.frame(h2o.getFrame(paste0("cv_holdout_prediction_", model_id))["penetrating"],
col.names = paste0(model_id, "_", length(base_models), "_models")
))
# ----------------------------------
# super learner base model reduction
# ----------------------------------
while (TRUE) {
meta <- h2o.getModel(paste0("metalearner_glm_superlearner_iter_", sl_iter, "_cv_1"))
names <- meta@model$coefficients_table[, "names"]
coeffs <- (meta@model$coefficients_table[, "standardized_coefficients"] > 0)
for (j in 2:nfolds) {
meta <- h2o.getModel(paste0("metalearner_glm_superlearner_iter_", sl_iter, "_cv_", j))
names <- meta@model$coefficients_table[, "names"]
coeffs <- coeffs + (meta@model$coefficients_table[, "standardized_coefficients"] > 0)
}
base_models_ <- as.list(names[coeffs >= ceiling(nfolds / 2) & names != "Intercept"])
if (length(base_models_) == 0) {
cat("No base models passing the threshold\n\n")
break
}
if (sum(base_models %in% base_models_) == length(base_models)) {
cat("No further reduction of base models\n\n")
break
}
sl_iter <- sl_iter + 1
base_models <- base_models_
cat(paste0("Super learner iteration ", sl_iter, " (", length(base_models), " models)\n"))
sl <- h2o.stackedEnsemble(
x = x, y = y, model_id = paste0("superlearner_iter_", sl_iter),
training_frame = tmp, seed = 1,
base_models = base_models,
metalearner_algorithm = "glm",
metalearner_nfolds = nfolds,
keep_levelone_frame = TRUE,
metalearner_params = list(
standardize = TRUE, keep_cross_validation_predictions = TRUE,
balance_classes = TRUE, class_sampling_factors = samp_factors
)
)
tmp_path <- h2o.saveModel(h2o.getModel(paste0("superlearner_iter_", sl_iter)), path = exp_dir, force = TRUE)
model_id <- paste0("metalearner_glm_superlearner_iter_", sl_iter)
tmp_path <- h2o.saveModel(h2o.getModel(model_id), path = exp_dir, force = TRUE)
res <- cbind(res, as.data.frame(h2o.getFrame(paste0("cv_holdout_prediction_", model_id))["penetrating"],
col.names = paste0(model_id, "_", length(base_models), "_models")
))
}
# -----------------------------------------
# super learner for homogeneous base models
# -----------------------------------------
# DeepLearning
base_models <- deeplearning_1@model_ids
cat(paste0("Super learner deep learning (", length(base_models), " models)\n"))
sl <- h2o.stackedEnsemble(
x = x, y = y, model_id = "superlearner_deeplearning",
training_frame = tmp, seed = 1,
base_models = base_models,
metalearner_algorithm = "glm",
metalearner_nfolds = nfolds,
keep_levelone_frame = TRUE,
metalearner_params = list(
standardize = TRUE, keep_cross_validation_predictions = TRUE,
balance_classes = TRUE, class_sampling_factors = samp_factors
)
)
tmp_path <- h2o.saveModel(h2o.getModel("superlearner_deeplearning"), path = exp_dir, force = TRUE)
model_id <- "metalearner_glm_superlearner_deeplearning"
tmp_path <- h2o.saveModel(h2o.getModel(model_id), path = exp_dir, force = TRUE)
res <- cbind(res, as.data.frame(h2o.getFrame(paste0("cv_holdout_prediction_", model_id))["penetrating"],
col.names = paste0(model_id, "_", length(base_models), "_models")
))
# GBM
base_models <- as.list(c(unlist(gbm@model_ids), paste0("GBM_", 1:5)))
cat(paste0("Super learner GBM (", length(base_models), " models)\n"))
sl <- h2o.stackedEnsemble(
x = x, y = y, model_id = "superlearner_gbm",
training_frame = tmp, seed = 1,
base_models = base_models,
metalearner_algorithm = "glm",
metalearner_nfolds = nfolds,
keep_levelone_frame = TRUE,
metalearner_params = list(
standardize = TRUE, keep_cross_validation_predictions = TRUE,
balance_classes = TRUE, class_sampling_factors = samp_factors
)
)
tmp_path <- h2o.saveModel(h2o.getModel("superlearner_gbm"), path = exp_dir, force = TRUE)
model_id <- "metalearner_glm_superlearner_gbm"
tmp_path <- h2o.saveModel(h2o.getModel(model_id), path = exp_dir, force = TRUE)
res <- cbind(res, as.data.frame(h2o.getFrame(paste0("cv_holdout_prediction_", model_id))["penetrating"],
col.names = paste0(model_id, "_", length(base_models), "_models")
))
# XGBoost
base_models <- as.list(c(unlist(xgboost@model_ids), paste0("XGBoost_", 1:3)))
cat(paste0("Super learner XGBoost (", length(base_models), " models)\n"))
sl <- h2o.stackedEnsemble(
x = x, y = y, model_id = "superlearner_xgboost",
training_frame = tmp, seed = 1,
base_models = base_models,
metalearner_algorithm = "glm",
metalearner_nfolds = nfolds,
keep_levelone_frame = TRUE,
metalearner_params = list(
standardize = TRUE, keep_cross_validation_predictions = TRUE,
balance_classes = TRUE, class_sampling_factors = samp_factors
)
)
tmp_path <- h2o.saveModel(h2o.getModel("superlearner_xgboost"), path = exp_dir, force = TRUE)
model_id <- "metalearner_glm_superlearner_xgboost"
tmp_path <- h2o.saveModel(h2o.getModel(model_id), path = exp_dir, force = TRUE)
res <- cbind(res, as.data.frame(h2o.getFrame(paste0("cv_holdout_prediction_", model_id))["penetrating"],
col.names = paste0(model_id, "_", length(base_models), "_models")
))
write.csv(res, file = paste0(exp_dir, "/cv_holdout_predictions.csv"), row.names = FALSE)
cat("\n\n")
h2o.removeAll()
}5.3.2 Inner cross-validation
load(file = "./rdata/var_reduct_cpp_train_test_splits_y_shuffled.RData")
for (i in 1:10) {
cat(paste0("Outer training set ", i, "\n"))
prefix <- paste0("outer_", i)
exp_dir <- paste0("/Users/renee/Downloads/cell_penetration_adversarial/", prefix)
dir.create(exp_dir)
train_cpp_models(train_set = outer_splits[[i]][["train"]], exp_dir = exp_dir, prefix = prefix)
}# Keep track of grid models
dl_grid <- list()
gbm_grid <- list()
xgboost_grid <- list()
dl_grid_params <- list()
gbm_grid_params <- list()
xgboost_grid_params <- list()
for (i in 1:10) {
cat(paste0("Outer training set ", i, "\n"))
prefix <- paste0("outer_", i)
dir <- paste0("/Users/renee/Downloads/cell_penetration_adversarial/", prefix)
files <- list.files(dir)
# Deep learning
dl <- files[str_detect(files, "DeepLearning_model")]
for (m in dl) {
model <- h2o.loadModel(paste0(dir, "/", m))
hs <- sha1(paste(c(
model@allparameters$epsilon,
model@allparameters$hidden,
model@allparameters$hidden_dropout_ratios,
model@allparameters$input_dropout_ratio,
model@allparameters$rho
), collapse = " "))
if (hs %in% names(dl_grid)) {
dl_grid[[hs]] <- c(dl_grid[[hs]], m)
} else {
dl_grid[[hs]] <- c(m)
dl_grid_params <- list.append(
dl_grid_params,
c(
"epsilon" = model@allparameters$epsilon,
"hidden" = paste0("[", paste(model@allparameters$hidden, collapse = ","), "]"),
"hidden_dropout_ratios" = paste0(
"[",
paste(model@allparameters$hidden_dropout_ratios,
collapse = ","
), "]"
),
"input_dropout_ratio" = model@allparameters$input_dropout_ratio,
"rho" = model@allparameters$rho
)
)
}
}
h2o.removeAll()
# GBM
gbm <- files[str_detect(files, "GBM_model")]
for (m in gbm) {
model <- h2o.loadModel(paste0(dir, "/", m))
hs <- sha1(paste(c(
model@allparameters$col_sample_rate,
model@allparameters$col_sample_rate_per_tree,
model@allparameters$max_depth,
model@allparameters$min_rows,
model@allparameters$min_split_improvement,
model@allparameters$sample_rate
), collapse = " "))
if (hs %in% names(gbm_grid)) {
gbm_grid[[hs]] <- c(gbm_grid[[hs]], m)
} else {
gbm_grid[[hs]] <- c(m)
gbm_grid_params <- list.append(
gbm_grid_params,
c(
"col_sample_rate" = model@allparameters$col_sample_rate,
"col_sample_rate_per_tree" = model@allparameters$col_sample_rate_per_tree,
"max_depth" = model@allparameters$max_depth,
"min_rows" = model@allparameters$min_rows,
"min_split_improvement" = model@allparameters$min_split_improvement,
"sample_rate" = model@allparameters$sample_rate
)
)
}
}
h2o.removeAll()
# XGBoost
xgboost <- files[str_detect(files, "XGBoost_model")]
for (m in xgboost) {
model <- h2o.loadModel(paste0(dir, "/", m))
hs <- sha1(paste(c(
model@allparameters$booster,
model@allparameters$col_sample_rate,
model@allparameters$col_sample_rate_per_tree,
model@allparameters$max_depth,
model@allparameters$min_rows,
model@allparameters$reg_alpha,
model@allparameters$reg_lambda,
model@allparameters$sample_rate
), collapse = " "))
if (hs %in% names(xgboost_grid)) {
xgboost_grid[[hs]] <- c(xgboost_grid[[hs]], m)
} else {
xgboost_grid[[hs]] <- c(m)
xgboost_grid_params <- list.append(
xgboost_grid_params,
c(
"booster" = model@allparameters$booster,
"col_sample_rate" = model@allparameters$col_sample_rate,
"col_sample_rate_per_tree" = model@allparameters$col_sample_rate_per_tree,
"max_depth" = model@allparameters$max_depth,
"min_rows" = model@allparameters$min_rows,
"reg_alpha" = model@allparameters$reg_alpha,
"reg_lambda" = model@allparameters$reg_lambda,
"sample_rate" = model@allparameters$sample_rate
)
)
}
}
h2o.removeAll()
}
dl_grid_params <- as.data.frame(t(data.frame(dl_grid_params)))
rownames(dl_grid_params) <- paste("Neural network grid model", 1:nrow(dl_grid_params))
gbm_grid_params <- as.data.frame(t(data.frame(gbm_grid_params)))
rownames(gbm_grid_params) <- paste("GBM grid model", 1:nrow(gbm_grid_params))
xgboost_grid_params <- as.data.frame(t(data.frame(xgboost_grid_params)))
rownames(xgboost_grid_params) <- paste("XGBoost grid model", 1:nrow(xgboost_grid_params))
write.csv(dl_grid_params, "./other_data/cell_penetration_adversarial/neural_network_grid_params.csv")
write.csv(gbm_grid_params, "./other_data/cell_penetration_adversarial/gbm_grid_params.csv")
write.csv(xgboost_grid_params, "./other_data/cell_penetration_adversarial/xgboost_grid_params.csv")
save(dl_grid, gbm_grid, xgboost_grid, file = "./rdata/model_training_grid_models_cpp_ad.RData")5.3.3 Model evaluation
model_evaluation <- function(holdout_pred, fold_asign, grid_name, grid_meta = NULL) {
res_ <- list()
model_num <- c()
if (startsWith(grid_name, "Super learner")) {
if (grid_name == "Super learner all models") {
m <- colnames(holdout_pred)[startsWith(colnames(holdout_pred), "metalearner_glm_superlearner_iter_0")]
} else if (grid_name == "Super learner final") {
sl_models <- colnames(holdout_pred)[startsWith(colnames(holdout_pred), "metalearner_glm_superlearner_iter")]
m <- sl_models[length(sl_models)]
} else if (grid_name == "Super learner neural network") {
m <- colnames(holdout_pred)[startsWith(colnames(holdout_pred), "metalearner_glm_superlearner_deeplearning")]
} else if (grid_name == "Super learner GBM") {
m <- colnames(holdout_pred)[startsWith(colnames(holdout_pred), "metalearner_glm_superlearner_gbm")]
} else if (grid_name == "Super learner XGBoost") {
m <- colnames(holdout_pred)[startsWith(colnames(holdout_pred), "metalearner_glm_superlearner_xgboost")]
}
logloss <- c()
MCC <- c()
F1 <- c()
acc <- c()
EF <- c()
BEDROC <- c()
threshold <- 0.5
for (k in 1:10) {
y_true <- sapply(holdout_pred[fold_assign == k, "category"], function(x) if (x == "penetrating") 1 else 0)
y_pred <- holdout_pred[fold_assign == k, m]
y_pred_cls <- sapply(y_pred, function(x) if (x >= threshold) 1 else 0)
logloss <- c(logloss, LogLoss(y_pred = y_pred, y_true = y_true))
MCC <- c(MCC, mcc(preds = y_pred_cls, actuals = y_true))
F1 <- c(F1, F1_Score(y_pred = y_pred_cls, y_true = y_true))
acc <- c(acc, balanced_accuracy(y_pred = y_pred_cls, y_true = y_true))
EF <- c(EF, enrichment_factor(x = y_pred, y = y_true, top = 0.05))
BEDROC <- c(BEDROC, bedroc(x = y_pred, y = y_true, alpha = 20))
}
res_ <- list.append(res_, c(
mean(logloss, na.rm = TRUE), sem(logloss),
mean(MCC, na.rm = TRUE), sem(MCC),
mean(F1, na.rm = TRUE), sem(F1),
mean(acc, na.rm = TRUE), sem(acc),
mean(EF, na.rm = TRUE), sem(EF),
mean(BEDROC, na.rm = TRUE), sem(BEDROC),
logloss, MCC, F1, acc, EF, BEDROC
))
} else {
for (j in 1:length(grid_meta)) {
g <- grid_meta[[j]]
m <- intersect(colnames(holdout_pred), g)
if (length(m) == 1) {
model_num <- c(model_num, j)
logloss <- c()
MCC <- c()
F1 <- c()
acc <- c()
EF <- c()
BEDROC <- c()
threshold <- 0.5
for (k in 1:10) {
y_true <- sapply(holdout_pred[fold_assign == k, "category"], function(x) if (x == "penetrating") 1 else 0)
y_pred <- holdout_pred[fold_assign == k, m]
y_pred_cls <- sapply(y_pred, function(x) if (x >= threshold) 1 else 0)
logloss <- c(logloss, LogLoss(y_pred = y_pred, y_true = y_true))
MCC <- c(MCC, mcc(preds = y_pred_cls, actuals = y_true))
F1 <- c(F1, F1_Score(y_pred = y_pred_cls, y_true = y_true))
acc <- c(acc, balanced_accuracy(y_pred = y_pred_cls, y_true = y_true))
EF <- c(EF, enrichment_factor(x = y_pred, y = y_true, top = 0.05))
BEDROC <- c(BEDROC, bedroc(x = y_pred, y = y_true, alpha = 20))
}
res_ <- list.append(res_, c(
mean(logloss, na.rm = TRUE), sem(logloss),
mean(MCC, na.rm = TRUE), sem(MCC),
mean(F1, na.rm = TRUE), sem(F1),
mean(acc, na.rm = TRUE), sem(acc),
mean(EF, na.rm = TRUE), sem(EF),
mean(BEDROC, na.rm = TRUE), sem(BEDROC),
logloss, MCC, F1, acc, EF, BEDROC
))
}
}
}
res <- as.data.frame(t(data.frame(res_)))
colnames(res) <- c(
"Log loss mean", "Log loss s.e.m.", "MCC mean", "MCC s.e.m.", "F_1 mean", "F_1 s.e.m.",
"Balanced accuracy mean", "Balanced accuracy s.e.m.", "EF mean", "EF s.e.m.", "BEDROC mean", "BEDROC s.e.m.",
paste("Log loss CV", 1:10), paste("MCC CV", 1:10), paste("F_1 CV", 1:10),
paste("Balanced accuracy CV", 1:10), paste("EF CV", 1:10), paste("BEDROC CV", 1:10)
)
if (nrow(res) == 1) {
rownames(res) <- grid_name
} else {
rownames(res) <- paste(grid_name, model_num)
}
return(res)
}5.3.4 Inner loop model selection
load(file = "./rdata/model_training_grid_models_cpp_ad.RData")
for (i in 1:10) {
holdout_pred <- read.csv(paste0("./other_data/cell_penetration_adversarial/outer_", i, "/cv_holdout_predictions.csv"))
fold_assign <- rep(1:10, ceiling(nrow(holdout_pred) / 10))[1:nrow(holdout_pred)]
data_ <- list()
# Deep learning grid models
data_ <- list.append(data_, model_evaluation(holdout_pred, fold_assign,
grid_name = "Neural network grid model",
grid_meta = dl_grid
))
# GBM grid models
data_ <- list.append(data_, model_evaluation(holdout_pred, fold_assign,
grid_name = "GBM grid model",
grid_meta = gbm_grid
))
# GBM default models
data_ <- list.append(data_, model_evaluation(holdout_pred, fold_assign,
grid_name = "GBM model",
grid_meta = as.list(paste0("GBM_", 1:5))
))
# XGBoost grid models
data_ <- list.append(data_, model_evaluation(holdout_pred, fold_assign,
grid_name = "XGBoost grid model",
grid_meta = xgboost_grid
))
# XGBoost default models
data_ <- list.append(data_, model_evaluation(holdout_pred, fold_assign,
grid_name = "XGBoost model",
grid_meta = as.list(paste0("XGBoost_", 1:3))
))
# DRF
data_ <- list.append(data_, model_evaluation(holdout_pred, fold_assign, grid_name = "DRF model", grid_meta = list("DRF")))
# XRT
data_ <- list.append(data_, model_evaluation(holdout_pred, fold_assign, grid_name = "XRT model", grid_meta = list("XRT")))
# Super learner all
data_ <- list.append(data_, model_evaluation(holdout_pred, fold_assign, grid_name = "Super learner all models"))
# Super learner final
data_ <- list.append(data_, model_evaluation(holdout_pred, fold_assign, grid_name = "Super learner final"))
# Super learner deep learning
data_ <- list.append(data_, model_evaluation(holdout_pred, fold_assign, grid_name = "Super learner neural network"))
# Super learner GBM
data_ <- list.append(data_, model_evaluation(holdout_pred, fold_assign, grid_name = "Super learner GBM"))
# Super learner XGBoost
data_ <- list.append(data_, model_evaluation(holdout_pred, fold_assign, grid_name = "Super learner XGBoost"))
data <- do.call(rbind, data_)
write.csv(data, paste0("./other_data/cell_penetration_adversarial/outer_", i, "/cv_res.csv"))
}5.3.5 Final evaluation
load(file = "./rdata/var_reduct_cpp_train_test_splits_y_shuffled.RData")
load(file = "./rdata/model_training_grid_models_cpp_ad.RData")
dir <- "/Users/renee/Downloads/cell_penetration_adversarial"
selected_models <- c(
"GBM grid model 68", "Super learner XGBoost", "GBM grid model 75",
"Super learner XGBoost", "GBM grid model 21", "Super learner final",
"GBM grid model 15", "GBM grid model 6", "GBM grid model 76",
"Neural network grid model 86"
)
logloss <- c()
MCC <- c()
F1 <- c()
acc <- c()
EF <- c()
BEDROC <- c()
threshold <- 0.5
for (i in 1:10) {
holdout_pred <- read.csv(paste0("./other_data/cell_penetration_adversarial/outer_", i, "/cv_holdout_predictions.csv"))
if (startsWith(selected_models[i], "Neural network grid model")) {
grid_num <- as.integer(str_extract(selected_models[i], "[0-9]+"))
m <- intersect(colnames(holdout_pred), dl_grid[[grid_num]])
model <- h2o.loadModel(paste0(dir, "/outer_", i, "/", m))
} else if (startsWith(selected_models[i], "GBM grid model")) {
grid_num <- as.integer(str_extract(selected_models[i], "[0-9]+"))
m <- intersect(colnames(holdout_pred), gbm_grid[[grid_num]])
model <- h2o.loadModel(paste0(dir, "/outer_", i, "/", m))
} else if (startsWith(selected_models[i], "GBM model")) {
grid_num <- as.integer(str_extract(selected_models[i], "[0-9]+"))
model <- h2o.loadModel(paste0(dir, "/outer_", i, "/GBM_", grid_num))
} else if (startsWith(selected_models[i], "XGBoost grid model")) {
grid_num <- as.integer(str_extract(selected_models[i], "[0-9]+"))
m <- intersect(colnames(holdout_pred), xgboost_grid[[grid_num]])
model <- h2o.loadModel(paste0(dir, "/outer_", i, "/", m))
} else if (startsWith(selected_models[i], "XGBoost model")) {
grid_num <- as.integer(str_extract(selected_models[i], "[0-9]+"))
model <- h2o.loadModel(paste0(dir, "/outer_", i, "/XGBoost_", grid_num))
} else if (selected_models[i] == "Super learner all models") {
model <- h2o.loadModel(paste0(dir, "/outer_", i, "/superlearner_iter_0"))
} else if (selected_models[i] == "Super learner final") {
files <- list.files(paste0(dir, "/outer_", i))
files <- files[startsWith(files, "superlearner_iter")]
model <- h2o.loadModel(paste0(dir, "/outer_", i, "/", files[length(files)]))
} else if (selected_models[i] == "Super learner neural network") {
model <- h2o.loadModel(paste0(dir, "/outer_", i, "/superlearner_deeplearning"))
} else if (selected_models[i] == "Super learner GBM") {
model <- h2o.loadModel(paste0(dir, "/outer_", i, "/superlearner_gbm"))
} else if (selected_models[i] == "Super learner XGBoost") {
model <- h2o.loadModel(paste0(dir, "/outer_", i, "/superlearner_xgboost"))
} else {
model <- h2o.loadModel(paste0(dir, "/outer_", i, "/", unlist(strsplit(selected_models[i], " "))[1]))
}
tmp <- as.h2o(subset(outer_splits[[i]][["test"]], select = -category))
y_true <- sapply(outer_splits[[i]][["test"]]$category, function(x) if (x == "penetrating") 1 else 0)
y_pred <- as.data.frame(as.data.frame(h2o.predict(model, tmp)))[, "penetrating"]
y_pred_cls <- sapply(y_pred, function(x) if (x >= threshold) 1 else 0)
logloss <- c(logloss, LogLoss(y_pred = y_pred, y_true = y_true))
MCC <- c(MCC, mcc(preds = y_pred_cls, actuals = y_true))
F1 <- c(F1, F1_Score(y_pred = y_pred_cls, y_true = y_true))
acc <- c(acc, balanced_accuracy(y_pred = y_pred_cls, y_true = y_true))
EF <- c(EF, enrichment_factor(x = y_pred, y = y_true, top = 0.05))
BEDROC <- c(BEDROC, bedroc(x = y_pred, y = y_true, alpha = 20))
}
save(logloss, MCC, F1, acc, EF, BEDROC, file = "./rdata/final_evaluation_cpp_ad.RData")load(file = "./rdata/final_evaluation_cpp_ad.RData")
data.frame(
Metric = c("Log loss", "MCC", "F<sub>1</sub>", "Balanced accuracy", "EF", "BEDROC"),
`Mean ± s.e.m.` = c(
paste0(round(mean(logloss), 3), " ± ", round(sem(logloss), 3)),
paste0(round(mean(MCC), 3), " ± ", round(sem(MCC), 3)),
paste0(round(mean(F1), 3), " ± ", round(sem(F1), 3)),
paste0(round(mean(acc), 3), " ± ", round(sem(acc), 3)),
paste0(round(mean(EF), 3), " ± ", round(sem(EF), 3)),
paste0(round(mean(BEDROC), 3), " ± ", round(sem(BEDROC), 3))
),
check.names = FALSE
) %>%
datatable(escape = FALSE, rownames = FALSE)5.4 Toxicity (classification)
load(file = "./rdata/var_reduct_tx_train_test_splits.RData")
set.seed(32)
# Shuffle the response variable of cross-validation training sets
for (i in 1:10) {
response <- outer_splits[[i]]$train$category
response <- sample(response)
outer_splits[[i]]$train$category <- response
}
# Shuffle the response variable of the whole data set
response <- tx_data$category
response <- sample(response)
tx_data$category <- response
save(tx_data, outer_splits,
file = "./rdata/var_reduct_tx_train_test_splits_y_shuffled.RData"
)5.4.1 Model training
train_tx_models <- function(train_set, exp_dir, prefix, nfolds = 10, grid_seed = 1) {
tmp <- as.h2o(train_set, destination_frame = prefix)
tmp["category"] <- as.factor(tmp["category"])
y <- "category"
x <- setdiff(names(tmp), y)
res <- as.data.frame(tmp$category)
samp_factors <- as.vector(mean(table(train_set$category)) / table(train_set$category))
# -------------------
# base model training
# -------------------
cat("Deep learning grid 1\n")
deeplearning_1 <- h2o.grid(
algorithm = "deeplearning", x = x, y = y, training_frame = tmp, seed = 1, nfolds = nfolds,
keep_cross_validation_predictions = TRUE, hyper_params = deeplearning_params_1,
stopping_rounds = 3, balance_classes = TRUE, class_sampling_factors = samp_factors,
max_after_balance_size = 0.5,
search_criteria = list(
strategy = "RandomDiscrete",
max_models = 100, seed = grid_seed
),
keep_cross_validation_models = FALSE, fold_assignment = "Modulo", parallelism = 0
)
for (model_id in deeplearning_1@model_ids) {
tmp_path <- h2o.saveModel(h2o.getModel(model_id), path = exp_dir, force = TRUE)
}
cat("GBM grid\n")
gbm <- h2o.grid(
algorithm = "gbm", x = x, y = y, training_frame = tmp, seed = 1, nfolds = nfolds,
keep_cross_validation_predictions = TRUE, hyper_params = gbm_params, stopping_rounds = 3,
balance_classes = TRUE, class_sampling_factors = samp_factors, max_after_balance_size = 0.5,
search_criteria = list(strategy = "RandomDiscrete", max_models = 100, seed = grid_seed),
keep_cross_validation_models = FALSE, fold_assignment = "Modulo", parallelism = 0
)
for (model_id in gbm@model_ids) {
tmp_path <- h2o.saveModel(h2o.getModel(model_id), path = exp_dir, force = TRUE)
}
cat("GBM 5 default models\n")
gbm_1 <- h2o.gbm(
model_id = "GBM_1", x = x, y = y, training_frame = tmp, seed = 1, nfolds = nfolds,
keep_cross_validation_predictions = TRUE, stopping_rounds = 3, score_tree_interval = 5,
balance_classes = TRUE, class_sampling_factors = samp_factors, max_after_balance_size = 0.5,
col_sample_rate = 0.8, col_sample_rate_per_tree = 0.8, learn_rate = 0.1,
max_depth = 6, min_rows = 1, min_split_improvement = 1e-5, ntrees = 10000, sample_rate = 0.8,
keep_cross_validation_models = FALSE, fold_assignment = "Modulo"
)
gbm_2 <- h2o.gbm(
model_id = "GBM_2", x = x, y = y, training_frame = tmp, seed = 1, nfolds = nfolds,
keep_cross_validation_predictions = TRUE, stopping_rounds = 3, score_tree_interval = 5,
balance_classes = TRUE, class_sampling_factors = samp_factors, max_after_balance_size = 0.5,
col_sample_rate = 0.8, col_sample_rate_per_tree = 0.8, learn_rate = 0.1,
max_depth = 7, min_rows = 10, min_split_improvement = 1e-5, ntrees = 10000, sample_rate = 0.8,
keep_cross_validation_models = FALSE, fold_assignment = "Modulo"
)
gbm_3 <- h2o.gbm(
model_id = "GBM_3", x = x, y = y, training_frame = tmp, seed = 1, nfolds = nfolds,
keep_cross_validation_predictions = TRUE, stopping_rounds = 3, score_tree_interval = 5,
balance_classes = TRUE, class_sampling_factors = samp_factors, max_after_balance_size = 0.5,
col_sample_rate = 0.8, col_sample_rate_per_tree = 0.8, learn_rate = 0.1,
max_depth = 8, min_rows = 10, min_split_improvement = 1e-5, ntrees = 10000, sample_rate = 0.8,
keep_cross_validation_models = FALSE, fold_assignment = "Modulo"
)
gbm_4 <- h2o.gbm(
model_id = "GBM_4", x = x, y = y, training_frame = tmp, seed = 1, nfolds = nfolds,
keep_cross_validation_predictions = TRUE, stopping_rounds = 3, score_tree_interval = 5,
balance_classes = TRUE, class_sampling_factors = samp_factors, max_after_balance_size = 0.5,
col_sample_rate = 0.8, col_sample_rate_per_tree = 0.8, learn_rate = 0.1,
max_depth = 10, min_rows = 10, min_split_improvement = 1e-5, ntrees = 10000, sample_rate = 0.8,
keep_cross_validation_models = FALSE, fold_assignment = "Modulo"
)
gbm_5 <- h2o.gbm(
model_id = "GBM_5", x = x, y = y, training_frame = tmp, seed = 1, nfolds = nfolds,
keep_cross_validation_predictions = TRUE, stopping_rounds = 3, score_tree_interval = 5,
balance_classes = TRUE, class_sampling_factors = samp_factors, max_after_balance_size = 0.5,
col_sample_rate = 0.8, col_sample_rate_per_tree = 0.8, learn_rate = 0.1,
max_depth = 15, min_rows = 100, min_split_improvement = 1e-5, ntrees = 10000, sample_rate = 0.8,
keep_cross_validation_models = FALSE, fold_assignment = "Modulo"
)
for (model_id in paste0("GBM_", 1:5)) {
tmp_path <- h2o.saveModel(h2o.getModel(model_id), path = exp_dir, force = TRUE)
}
cat("XGBoost grid\n")
xgboost <- h2o.grid(
algorithm = "xgboost", x = x, y = y, training_frame = tmp, seed = 1, nfolds = nfolds,
keep_cross_validation_predictions = TRUE, hyper_params = xgboost_params, stopping_rounds = 3,
search_criteria = list(strategy = "RandomDiscrete", max_models = 100, seed = grid_seed),
keep_cross_validation_models = FALSE, fold_assignment = "Modulo", parallelism = 0
)
for (model_id in xgboost@model_ids) {
tmp_path <- h2o.saveModel(h2o.getModel(model_id), path = exp_dir, force = TRUE)
}
cat("XGBoost 3 default models\n")
xgboost_1 <- h2o.xgboost(
model_id = "XGBoost_1", x = x, y = y, training_frame = tmp, seed = 1, nfolds = nfolds,
keep_cross_validation_predictions = TRUE, stopping_rounds = 3, score_tree_interval = 5,
booster = "gbtree", col_sample_rate = 0.8, col_sample_rate_per_tree = 0.8,
max_depth = 10, min_rows = 5, ntrees = 10000, reg_alpha = 0, reg_lambda = 1,
sample_rate = 0.6, keep_cross_validation_models = FALSE, fold_assignment = "Modulo"
)
xgboost_2 <- h2o.xgboost(
model_id = "XGBoost_2", x = x, y = y, training_frame = tmp, seed = 1, nfolds = nfolds,
keep_cross_validation_predictions = TRUE, stopping_rounds = 3, score_tree_interval = 5,
booster = "gbtree", col_sample_rate = 0.8, col_sample_rate_per_tree = 0.8,
max_depth = 20, min_rows = 10, ntrees = 10000, reg_alpha = 0, reg_lambda = 1,
sample_rate = 0.6, keep_cross_validation_models = FALSE, fold_assignment = "Modulo"
)
xgboost_3 <- h2o.xgboost(
model_id = "XGBoost_3", x = x, y = y, training_frame = tmp, seed = 1, nfolds = nfolds,
keep_cross_validation_predictions = TRUE, stopping_rounds = 3, score_tree_interval = 5,
booster = "gbtree", col_sample_rate = 0.8, col_sample_rate_per_tree = 0.8,
max_depth = 5, min_rows = 3, ntrees = 10000, reg_alpha = 0, reg_lambda = 1,
sample_rate = 0.8, keep_cross_validation_models = FALSE, fold_assignment = "Modulo"
)
for (model_id in paste0("XGBoost_", 1:3)) {
tmp_path <- h2o.saveModel(h2o.getModel(model_id), path = exp_dir, force = TRUE)
}
cat("GLM\n")
glm <- h2o.glm(
model_id = "GLM", x = x, y = y, training_frame = tmp, seed = 1, nfolds = nfolds,
keep_cross_validation_predictions = TRUE, alpha = c(0.0, 0.2, 0.4, 0.6, 0.8, 1.0),
balance_classes = TRUE, class_sampling_factors = samp_factors, max_after_balance_size = 0.5,
keep_cross_validation_models = FALSE, fold_assignment = "Modulo"
)
tmp_path <- h2o.saveModel(h2o.getModel("GLM"), path = exp_dir, force = TRUE)
cat("DRF\n")
drf <- h2o.randomForest(
model_id = "DRF", x = x, y = y, training_frame = tmp, seed = 1, nfolds = nfolds,
keep_cross_validation_predictions = TRUE, ntrees = 10000,
score_tree_interval = 5, stopping_rounds = 3,
balance_classes = TRUE, class_sampling_factors = samp_factors, max_after_balance_size = 0.5,
keep_cross_validation_models = FALSE, fold_assignment = "Modulo"
)
tmp_path <- h2o.saveModel(h2o.getModel("DRF"), path = exp_dir, force = TRUE)
cat("XRT\n")
xrt <- h2o.randomForest(
model_id = "XRT", x = x, y = y, training_frame = tmp, seed = 1, nfolds = nfolds,
keep_cross_validation_predictions = TRUE, ntrees = 10000, histogram_type = "Random",
score_tree_interval = 5, stopping_rounds = 3,
balance_classes = TRUE, class_sampling_factors = samp_factors, max_after_balance_size = 0.5,
keep_cross_validation_models = FALSE, fold_assignment = "Modulo"
)
tmp_path <- h2o.saveModel(h2o.getModel("XRT"), path = exp_dir, force = TRUE)
# -----------------------
# get holdout predictions
# -----------------------
base_models <- as.list(c(
unlist(deeplearning_1@model_ids),
unlist(gbm@model_ids), paste0("GBM_", 1:5),
unlist(xgboost@model_ids), paste0("XGBoost_", 1:3),
"GLM",
"DRF",
"XRT"
))
for (model_id in base_models) {
res <- cbind(res, as.data.frame(h2o.getFrame(paste0("cv_holdout_prediction_", model_id))[3],
col.names = model_id
))
}
# ----------------------
# super learner training
# ----------------------
sl_iter <- 0
cat(paste0("Super learner iteration ", sl_iter, " (", length(base_models), " models)\n"))
sl <- h2o.stackedEnsemble(
x = x, y = y, model_id = paste0("superlearner_iter_", sl_iter),
training_frame = tmp, seed = 1,
base_models = base_models,
metalearner_algorithm = "glm",
metalearner_nfolds = nfolds,
keep_levelone_frame = TRUE,
metalearner_params = list(
standardize = TRUE, keep_cross_validation_predictions = TRUE,
balance_classes = TRUE, class_sampling_factors = samp_factors,
max_after_balance_size = 0.5
)
)
tmp_path <- h2o.saveModel(h2o.getModel(paste0("superlearner_iter_", sl_iter)), path = exp_dir, force = TRUE)
model_id <- paste0("metalearner_glm_superlearner_iter_", sl_iter)
tmp_path <- h2o.saveModel(h2o.getModel(model_id), path = exp_dir, force = TRUE)
res <- cbind(res, as.data.frame(h2o.getFrame(paste0("cv_holdout_prediction_", model_id))[3],
col.names = paste0(model_id, "_", length(base_models), "_models")
))
# ----------------------------------
# super learner base model reduction
# ----------------------------------
while (TRUE) {
meta <- h2o.getModel(paste0("metalearner_glm_superlearner_iter_", sl_iter, "_cv_1"))
names <- meta@model$coefficients_table[, "names"]
coeffs <- (meta@model$coefficients_table[, "standardized_coefficients"] > 0)
for (j in 2:nfolds) {
meta <- h2o.getModel(paste0("metalearner_glm_superlearner_iter_", sl_iter, "_cv_", j))
names <- meta@model$coefficients_table[, "names"]
coeffs <- coeffs + (meta@model$coefficients_table[, "standardized_coefficients"] > 0)
}
base_models_ <- as.list(names[coeffs >= ceiling(nfolds / 2) & names != "Intercept"])
if (length(base_models_) == 0) {
cat("No base models passing the threshold\n\n")
break
}
if (sum(base_models %in% base_models_) == length(base_models)) {
cat("No further reduction of base models\n\n")
break
}
sl_iter <- sl_iter + 1
base_models <- base_models_
cat(paste0("Super learner iteration ", sl_iter, " (", length(base_models), " models)\n"))
sl <- h2o.stackedEnsemble(
x = x, y = y, model_id = paste0("superlearner_iter_", sl_iter),
training_frame = tmp, seed = 1,
base_models = base_models,
metalearner_algorithm = "glm",
metalearner_nfolds = nfolds,
keep_levelone_frame = TRUE,
metalearner_params = list(
standardize = TRUE, keep_cross_validation_predictions = TRUE,
balance_classes = TRUE, class_sampling_factors = samp_factors,
max_after_balance_size = 0.5
)
)
tmp_path <- h2o.saveModel(h2o.getModel(paste0("superlearner_iter_", sl_iter)), path = exp_dir, force = TRUE)
model_id <- paste0("metalearner_glm_superlearner_iter_", sl_iter)
tmp_path <- h2o.saveModel(h2o.getModel(model_id), path = exp_dir, force = TRUE)
res <- cbind(res, as.data.frame(h2o.getFrame(paste0("cv_holdout_prediction_", model_id))[3],
col.names = paste0(model_id, "_", length(base_models), "_models")
))
}
# -----------------------------------------
# super learner for homogeneous base models
# -----------------------------------------
# DeepLearning
base_models <- deeplearning_1@model_ids
cat(paste0("Super learner deep learning (", length(base_models), " models)\n"))
sl <- h2o.stackedEnsemble(
x = x, y = y, model_id = "superlearner_deeplearning",
training_frame = tmp, seed = 1,
base_models = base_models,
metalearner_algorithm = "glm",
metalearner_nfolds = nfolds,
keep_levelone_frame = TRUE,
metalearner_params = list(
standardize = TRUE, keep_cross_validation_predictions = TRUE,
balance_classes = TRUE, class_sampling_factors = samp_factors,
max_after_balance_size = 0.5
)
)
tmp_path <- h2o.saveModel(h2o.getModel("superlearner_deeplearning"), path = exp_dir, force = TRUE)
model_id <- "metalearner_glm_superlearner_deeplearning"
tmp_path <- h2o.saveModel(h2o.getModel(model_id), path = exp_dir, force = TRUE)
res <- cbind(res, as.data.frame(h2o.getFrame(paste0("cv_holdout_prediction_", model_id))[3],
col.names = paste0(model_id, "_", length(base_models), "_models")
))
# GBM
base_models <- as.list(c(unlist(gbm@model_ids), paste0("GBM_", 1:5)))
cat(paste0("Super learner GBM (", length(base_models), " models)\n"))
sl <- h2o.stackedEnsemble(
x = x, y = y, model_id = "superlearner_gbm",
training_frame = tmp, seed = 1,
base_models = base_models,
metalearner_algorithm = "glm",
metalearner_nfolds = nfolds,
keep_levelone_frame = TRUE,
metalearner_params = list(
standardize = TRUE, keep_cross_validation_predictions = TRUE,
balance_classes = TRUE, class_sampling_factors = samp_factors,
max_after_balance_size = 0.5
)
)
tmp_path <- h2o.saveModel(h2o.getModel("superlearner_gbm"), path = exp_dir, force = TRUE)
model_id <- "metalearner_glm_superlearner_gbm"
tmp_path <- h2o.saveModel(h2o.getModel(model_id), path = exp_dir, force = TRUE)
res <- cbind(res, as.data.frame(h2o.getFrame(paste0("cv_holdout_prediction_", model_id))[3],
col.names = paste0(model_id, "_", length(base_models), "_models")
))
# XGBoost
base_models <- as.list(c(unlist(xgboost@model_ids), paste0("XGBoost_", 1:3)))
cat(paste0("Super learner XGBoost (", length(base_models), " models)\n"))
sl <- h2o.stackedEnsemble(
x = x, y = y, model_id = "superlearner_xgboost",
training_frame = tmp, seed = 1,
base_models = base_models,
metalearner_algorithm = "glm",
metalearner_nfolds = nfolds,
keep_levelone_frame = TRUE,
metalearner_params = list(
standardize = TRUE, keep_cross_validation_predictions = TRUE,
balance_classes = TRUE, class_sampling_factors = samp_factors,
max_after_balance_size = 0.5
)
)
tmp_path <- h2o.saveModel(h2o.getModel("superlearner_xgboost"), path = exp_dir, force = TRUE)
model_id <- "metalearner_glm_superlearner_xgboost"
tmp_path <- h2o.saveModel(h2o.getModel(model_id), path = exp_dir, force = TRUE)
res <- cbind(res, as.data.frame(h2o.getFrame(paste0("cv_holdout_prediction_", model_id))[3],
col.names = paste0(model_id, "_", length(base_models), "_models")
))
write.csv(res, file = paste0(exp_dir, "/cv_holdout_predictions.csv"), row.names = FALSE)
cat("\n\n")
h2o.removeAll()
}5.4.2 Inner cross-validation
load(file = "./rdata/var_reduct_tx_train_test_splits_y_shuffled.RData")
for (i in 1:10) {
cat(paste0("Outer training set ", i, "\n"))
prefix <- paste0("outer_", i)
exp_dir <- paste0("/Users/renee/Downloads/toxicity_adversarial/", prefix)
dir.create(exp_dir)
train_tx_models(train_set = outer_splits[[i]][["train"]], exp_dir = exp_dir, prefix = prefix)
}# Keep track of grid models
dl_grid <- list()
gbm_grid <- list()
xgboost_grid <- list()
dl_grid_params <- list()
gbm_grid_params <- list()
xgboost_grid_params <- list()
for (i in 1:10) {
cat(paste0("Outer training set ", i, "\n"))
prefix <- paste0("outer_", i)
dir <- paste0("/Users/renee/Downloads/toxicity_adversarial/", prefix)
files <- list.files(dir)
# Deep learning
dl <- files[str_detect(files, "DeepLearning_model")]
for (m in dl) {
model <- h2o.loadModel(paste0(dir, "/", m))
hs <- sha1(paste(c(
model@allparameters$epsilon,
model@allparameters$hidden,
model@allparameters$hidden_dropout_ratios,
model@allparameters$input_dropout_ratio,
model@allparameters$rho
), collapse = " "))
if (hs %in% names(dl_grid)) {
dl_grid[[hs]] <- c(dl_grid[[hs]], m)
} else {
dl_grid[[hs]] <- c(m)
dl_grid_params <- list.append(
dl_grid_params,
c(
"epsilon" = model@allparameters$epsilon,
"hidden" = paste0("[", paste(model@allparameters$hidden, collapse = ","), "]"),
"hidden_dropout_ratios" = paste0(
"[",
paste(model@allparameters$hidden_dropout_ratios,
collapse = ","
), "]"
),
"input_dropout_ratio" = model@allparameters$input_dropout_ratio,
"rho" = model@allparameters$rho
)
)
}
}
h2o.removeAll()
# GBM
gbm <- files[str_detect(files, "GBM_model")]
for (m in gbm) {
model <- h2o.loadModel(paste0(dir, "/", m))
hs <- sha1(paste(c(
model@allparameters$col_sample_rate,
model@allparameters$col_sample_rate_per_tree,
model@allparameters$max_depth,
model@allparameters$min_rows,
model@allparameters$min_split_improvement,
model@allparameters$sample_rate
), collapse = " "))
if (hs %in% names(gbm_grid)) {
gbm_grid[[hs]] <- c(gbm_grid[[hs]], m)
} else {
gbm_grid[[hs]] <- c(m)
gbm_grid_params <- list.append(
gbm_grid_params,
c(
"col_sample_rate" = model@allparameters$col_sample_rate,
"col_sample_rate_per_tree" = model@allparameters$col_sample_rate_per_tree,
"max_depth" = model@allparameters$max_depth,
"min_rows" = model@allparameters$min_rows,
"min_split_improvement" = model@allparameters$min_split_improvement,
"sample_rate" = model@allparameters$sample_rate
)
)
}
}
h2o.removeAll()
# XGBoost
xgboost <- files[str_detect(files, "XGBoost_model")]
for (m in xgboost) {
model <- h2o.loadModel(paste0(dir, "/", m))
hs <- sha1(paste(c(
model@allparameters$booster,
model@allparameters$col_sample_rate,
model@allparameters$col_sample_rate_per_tree,
model@allparameters$max_depth,
model@allparameters$min_rows,
model@allparameters$reg_alpha,
model@allparameters$reg_lambda,
model@allparameters$sample_rate
), collapse = " "))
if (hs %in% names(xgboost_grid)) {
xgboost_grid[[hs]] <- c(xgboost_grid[[hs]], m)
} else {
xgboost_grid[[hs]] <- c(m)
xgboost_grid_params <- list.append(
xgboost_grid_params,
c(
"booster" = model@allparameters$booster,
"col_sample_rate" = model@allparameters$col_sample_rate,
"col_sample_rate_per_tree" = model@allparameters$col_sample_rate_per_tree,
"max_depth" = model@allparameters$max_depth,
"min_rows" = model@allparameters$min_rows,
"reg_alpha" = model@allparameters$reg_alpha,
"reg_lambda" = model@allparameters$reg_lambda,
"sample_rate" = model@allparameters$sample_rate
)
)
}
}
h2o.removeAll()
}
dl_grid_params <- as.data.frame(t(data.frame(dl_grid_params)))
rownames(dl_grid_params) <- paste("Neural network grid model", 1:nrow(dl_grid_params))
gbm_grid_params <- as.data.frame(t(data.frame(gbm_grid_params)))
rownames(gbm_grid_params) <- paste("GBM grid model", 1:nrow(gbm_grid_params))
xgboost_grid_params <- as.data.frame(t(data.frame(xgboost_grid_params)))
rownames(xgboost_grid_params) <- paste("XGBoost grid model", 1:nrow(xgboost_grid_params))
write.csv(dl_grid_params, "./other_data/toxicity_adversarial/neural_network_grid_params.csv")
write.csv(gbm_grid_params, "./other_data/toxicity_adversarial/gbm_grid_params.csv")
write.csv(xgboost_grid_params, "./other_data/toxicity_adversarial/xgboost_grid_params.csv")
save(dl_grid, gbm_grid, xgboost_grid, file = "./rdata/model_training_grid_models_tx_ad.RData")5.4.3 Model evaluation
model_evaluation <- function(holdout_pred, fold_asign, grid_name, grid_meta = NULL) {
res_ <- list()
model_num <- c()
if (startsWith(grid_name, "Super learner")) {
if (grid_name == "Super learner all models") {
m <- colnames(holdout_pred)[startsWith(colnames(holdout_pred), "metalearner_glm_superlearner_iter_0")]
} else if (grid_name == "Super learner final") {
sl_models <- colnames(holdout_pred)[startsWith(colnames(holdout_pred), "metalearner_glm_superlearner_iter")]
m <- sl_models[length(sl_models)]
} else if (grid_name == "Super learner neural network") {
m <- colnames(holdout_pred)[startsWith(colnames(holdout_pred), "metalearner_glm_superlearner_deeplearning")]
} else if (grid_name == "Super learner GBM") {
m <- colnames(holdout_pred)[startsWith(colnames(holdout_pred), "metalearner_glm_superlearner_gbm")]
} else if (grid_name == "Super learner XGBoost") {
m <- colnames(holdout_pred)[startsWith(colnames(holdout_pred), "metalearner_glm_superlearner_xgboost")]
}
logloss <- c()
MCC <- c()
F1 <- c()
acc <- c()
EF <- c()
BEDROC <- c()
threshold <- 0.5
for (k in 1:10) {
y_true <- holdout_pred[fold_assign == k, "category"]
y_pred <- holdout_pred[fold_assign == k, m]
y_pred_cls <- sapply(y_pred, function(x) if (x >= threshold) 1 else 0)
logloss <- c(logloss, LogLoss(y_pred = y_pred, y_true = y_true))
MCC <- c(MCC, mcc(preds = y_pred_cls, actuals = y_true))
F1 <- c(F1, F1_Score(y_pred = y_pred_cls, y_true = y_true))
acc <- c(acc, balanced_accuracy(y_pred = y_pred_cls, y_true = y_true))
EF <- c(EF, enrichment_factor(x = y_pred, y = y_true, top = 0.05))
BEDROC <- c(BEDROC, bedroc(x = y_pred, y = y_true, alpha = 20))
}
res_ <- list.append(res_, c(
mean(logloss, na.rm = TRUE), sem(logloss),
mean(MCC, na.rm = TRUE), sem(MCC),
mean(F1, na.rm = TRUE), sem(F1),
mean(acc, na.rm = TRUE), sem(acc),
mean(EF, na.rm = TRUE), sem(EF),
mean(BEDROC, na.rm = TRUE), sem(BEDROC),
logloss, MCC, F1, acc, EF, BEDROC
))
} else {
for (j in 1:length(grid_meta)) {
g <- grid_meta[[j]]
m <- intersect(colnames(holdout_pred), g)
if (length(m) == 1) {
model_num <- c(model_num, j)
logloss <- c()
MCC <- c()
F1 <- c()
acc <- c()
EF <- c()
BEDROC <- c()
threshold <- 0.5
for (k in 1:10) {
y_true <- holdout_pred[fold_assign == k, "category"]
y_pred <- holdout_pred[fold_assign == k, m]
y_pred_cls <- sapply(y_pred, function(x) if (x >= threshold) 1 else 0)
logloss <- c(logloss, LogLoss(y_pred = y_pred, y_true = y_true))
MCC <- c(MCC, mcc(preds = y_pred_cls, actuals = y_true))
F1 <- c(F1, F1_Score(y_pred = y_pred_cls, y_true = y_true))
acc <- c(acc, balanced_accuracy(y_pred = y_pred_cls, y_true = y_true))
EF <- c(EF, enrichment_factor(x = y_pred, y = y_true, top = 0.05))
BEDROC <- c(BEDROC, bedroc(x = y_pred, y = y_true, alpha = 20))
}
res_ <- list.append(res_, c(
mean(logloss, na.rm = TRUE), sem(logloss),
mean(MCC, na.rm = TRUE), sem(MCC),
mean(F1, na.rm = TRUE), sem(F1),
mean(acc, na.rm = TRUE), sem(acc),
mean(EF, na.rm = TRUE), sem(EF),
mean(BEDROC, na.rm = TRUE), sem(BEDROC),
logloss, MCC, F1, acc, EF, BEDROC
))
}
}
}
res <- as.data.frame(t(data.frame(res_)))
colnames(res) <- c(
"Log loss mean", "Log loss s.e.m.", "MCC mean", "MCC s.e.m.", "F_1 mean", "F_1 s.e.m.",
"Balanced accuracy mean", "Balanced accuracy s.e.m.", "EF mean", "EF s.e.m.", "BEDROC mean", "BEDROC s.e.m.",
paste("Log loss CV", 1:10), paste("MCC CV", 1:10), paste("F_1 CV", 1:10),
paste("Balanced accuracy CV", 1:10), paste("EF CV", 1:10), paste("BEDROC CV", 1:10)
)
if (nrow(res) == 1) {
rownames(res) <- grid_name
} else {
rownames(res) <- paste(grid_name, model_num)
}
return(res)
}5.4.4 Inner loop model selection
load(file = "./rdata/model_training_grid_models_tx_ad.RData")
for (i in 1:10) {
holdout_pred <- read.csv(paste0("./other_data/toxicity_adversarial/outer_", i, "/cv_holdout_predictions.csv"))
fold_assign <- rep(1:10, ceiling(nrow(holdout_pred) / 10))[1:nrow(holdout_pred)]
data_ <- list()
# Deep learning grid models
data_ <- list.append(data_, model_evaluation(holdout_pred, fold_assign,
grid_name = "Neural network grid model",
grid_meta = dl_grid
))
# GBM grid models
data_ <- list.append(data_, model_evaluation(holdout_pred, fold_assign,
grid_name = "GBM grid model",
grid_meta = gbm_grid
))
# GBM default models
data_ <- list.append(data_, model_evaluation(holdout_pred, fold_assign,
grid_name = "GBM model",
grid_meta = as.list(paste0("GBM_", 1:5))
))
# XGBoost grid models
data_ <- list.append(data_, model_evaluation(holdout_pred, fold_assign,
grid_name = "XGBoost grid model",
grid_meta = xgboost_grid
))
# XGBoost default models
data_ <- list.append(data_, model_evaluation(holdout_pred, fold_assign,
grid_name = "XGBoost model",
grid_meta = as.list(paste0("XGBoost_", 1:3))
))
# GLM
data_ <- list.append(data_, model_evaluation(holdout_pred, fold_assign, grid_name = "GLM model", grid_meta = list("GLM")))
# DRF
data_ <- list.append(data_, model_evaluation(holdout_pred, fold_assign, grid_name = "DRF model", grid_meta = list("DRF")))
# XRT
data_ <- list.append(data_, model_evaluation(holdout_pred, fold_assign, grid_name = "XRT model", grid_meta = list("XRT")))
# Super learner all
data_ <- list.append(data_, model_evaluation(holdout_pred, fold_assign, grid_name = "Super learner all models"))
# Super learner final
data_ <- list.append(data_, model_evaluation(holdout_pred, fold_assign, grid_name = "Super learner final"))
# Super learner deep learning
data_ <- list.append(data_, model_evaluation(holdout_pred, fold_assign, grid_name = "Super learner neural network"))
# Super learner GBM
data_ <- list.append(data_, model_evaluation(holdout_pred, fold_assign, grid_name = "Super learner GBM"))
# Super learner XGBoost
data_ <- list.append(data_, model_evaluation(holdout_pred, fold_assign, grid_name = "Super learner XGBoost"))
data <- do.call(rbind, data_)
write.csv(data, paste0("./other_data/toxicity_adversarial/outer_", i, "/cv_res.csv"))
}5.4.5 Final evaluation
load(file = "./rdata/var_reduct_tx_train_test_splits_y_shuffled.RData")
load(file = "./rdata/model_training_grid_models_tx_ad.RData")
dir <- "/Users/renee/Downloads/toxicity_adversarial"
selected_models <- c(
"GBM grid model 52", "GBM grid model 95", "GBM grid model 48",
"GBM grid model 98", "GBM grid model 55", "GBM grid model 21",
"GBM grid model 23", "GBM grid model 54", "GBM grid model 3",
"GBM grid model 68"
)
logloss <- c()
MCC <- c()
F1 <- c()
acc <- c()
EF <- c()
BEDROC <- c()
threshold <- 0.5
for (i in 1:10) {
holdout_pred <- read.csv(paste0("./other_data/toxicity_adversarial/outer_", i, "/cv_holdout_predictions.csv"))
if (startsWith(selected_models[i], "Neural network grid model")) {
grid_num <- as.integer(str_extract(selected_models[i], "[0-9]+"))
m <- intersect(colnames(holdout_pred), dl_grid[[grid_num]])
model <- h2o.loadModel(paste0(dir, "/outer_", i, "/", m))
} else if (startsWith(selected_models[i], "GBM grid model")) {
grid_num <- as.integer(str_extract(selected_models[i], "[0-9]+"))
m <- intersect(colnames(holdout_pred), gbm_grid[[grid_num]])
model <- h2o.loadModel(paste0(dir, "/outer_", i, "/", m))
} else if (startsWith(selected_models[i], "GBM model")) {
grid_num <- as.integer(str_extract(selected_models[i], "[0-9]+"))
model <- h2o.loadModel(paste0(dir, "/outer_", i, "/GBM_", grid_num))
} else if (startsWith(selected_models[i], "XGBoost grid model")) {
grid_num <- as.integer(str_extract(selected_models[i], "[0-9]+"))
m <- intersect(colnames(holdout_pred), xgboost_grid[[grid_num]])
model <- h2o.loadModel(paste0(dir, "/outer_", i, "/", m))
} else if (startsWith(selected_models[i], "XGBoost model")) {
grid_num <- as.integer(str_extract(selected_models[i], "[0-9]+"))
model <- h2o.loadModel(paste0(dir, "/outer_", i, "/XGBoost_", grid_num))
} else if (selected_models[i] == "Super learner all models") {
model <- h2o.loadModel(paste0(dir, "/outer_", i, "/superlearner_iter_0"))
} else if (selected_models[i] == "Super learner final") {
files <- list.files(paste0(dir, "/outer_", i))
files <- files[startsWith(files, "superlearner_iter")]
model <- h2o.loadModel(paste0(dir, "/outer_", i, "/", files[length(files)]))
} else if (selected_models[i] == "Super learner neural network") {
model <- h2o.loadModel(paste0(dir, "/outer_", i, "/superlearner_deeplearning"))
} else if (selected_models[i] == "Super learner GBM") {
model <- h2o.loadModel(paste0(dir, "/outer_", i, "/superlearner_gbm"))
} else if (selected_models[i] == "Super learner XGBoost") {
model <- h2o.loadModel(paste0(dir, "/outer_", i, "/superlearner_xgboost"))
} else {
model <- h2o.loadModel(paste0(dir, "/outer_", i, "/", unlist(strsplit(selected_models[i], " "))[1]))
}
tmp <- as.h2o(subset(outer_splits[[i]][["test"]], select = -category))
y_true <- as.numeric(outer_splits[[i]][["test"]]$category) - 1
y_pred <- as.data.frame(as.data.frame(h2o.predict(model, tmp)))[, 3]
y_pred_cls <- sapply(y_pred, function(x) if (x >= threshold) 1 else 0)
logloss <- c(logloss, LogLoss(y_pred = y_pred, y_true = y_true))
MCC <- c(MCC, mcc(preds = y_pred_cls, actuals = y_true))
F1 <- c(F1, F1_Score(y_pred = y_pred_cls, y_true = y_true))
acc <- c(acc, balanced_accuracy(y_pred = y_pred_cls, y_true = y_true))
EF <- c(EF, enrichment_factor(x = y_pred, y = y_true, top = 0.05))
BEDROC <- c(BEDROC, bedroc(x = y_pred, y = y_true, alpha = 20))
}
save(logloss, MCC, F1, acc, EF, BEDROC, file = "./rdata/final_evaluation_tx_ad.RData")load(file = "./rdata/final_evaluation_tx_ad.RData")
data.frame(
Metric = c("Log loss", "MCC", "F<sub>1</sub>", "Balanced accuracy", "EF", "BEDROC"),
`Mean ± s.e.m.` = c(
paste0(round(mean(logloss), 3), " ± ", round(sem(logloss), 3)),
paste0(round(mean(MCC), 3), " ± ", round(sem(MCC), 3)),
paste0(round(mean(F1, na.rm = TRUE), 3), " ± ", round(sem(F1), 3)),
paste0(round(mean(acc), 3), " ± ", round(sem(acc), 3)),
paste0(round(mean(EF), 3), " ± ", round(sem(EF), 3)),
paste0(round(mean(BEDROC), 3), " ± ", round(sem(BEDROC), 3))
),
check.names = FALSE
) %>%
datatable(escape = FALSE, rownames = FALSE)5.5 Overall model interpretation
import h2o
import pandas as pd
import numpy as np
import shap
import matplotlib.pyplot as plt
import session_info5.5.1 Melanin binding (regression)
# Save train and test sets in csv format
load(file = "./rdata/var_reduct_mb_train_test_splits_y_shuffled.RData")
for (i in 1:10) {
prefix <- paste0("outer_", i)
exp_dir <- paste0("/Users/renee/Downloads/melanin_binding_adversarial/", prefix)
write.csv(outer_splits[[i]]$train, file = paste0(exp_dir, "/train_set.csv"))
write.csv(outer_splits[[i]]$test, file = paste0(exp_dir, "/test_set.csv"))
}h2o.init(nthreads=-1)
dir_path = '/Users/renee/Downloads/melanin_binding_adversarial'
model_names = ['superlearner_iter_6', 'superlearner_iter_5', 'superlearner_iter_6', 'superlearner_iter_6',
'superlearner_iter_7', 'superlearner_iter_7', 'superlearner_iter_6', 'superlearner_iter_7',
'superlearner_iter_7', 'superlearner_iter_6']
shap_res = []
for i in range(10):
print('Iter ' + str(i + 1) + ':')
train = pd.read_csv(dir_path + '/outer_' + str(i + 1) + '/train_set.csv', index_col=0)
X_train = train.iloc[:, :-1]
test = pd.read_csv(dir_path + '/outer_' + str(i + 1) + '/test_set.csv', index_col=0)
X_test = test.iloc[:, :-1]
model = h2o.load_model(dir_path + '/outer_' + str(i + 1) + '/' + model_names[i])
def model_predict_(data):
data = pd.DataFrame(data, columns=X_train.columns)
h2o_data = h2o.H2OFrame(data)
res = model.predict(h2o_data)
res = res.as_data_frame()
return(res)
np.random.seed(1)
X_train_ = X_train.iloc[np.random.choice(X_train.shape[0], 100, replace=False), :]
explainer = shap.KernelExplainer(model_predict_, X_train_, link='identity')
shap_values = explainer.shap_values(X_test, nsamples=1000)
shap_res.append(shap_values)
h2o.remove_all()
shap_res = pd.DataFrame(np.vstack(shap_res), columns=X_train.columns)
shap_res.to_csv('./other_data/mb_ad_shap_values_cv_data_sets.csv', index=False)load(file = "./rdata/var_reduct_mb_train_test_splits_y_shuffled.RData")
data <- do.call(rbind, lapply(outer_splits, function(x) x$test[-ncol(mb_data)]))
data <- apply(data, 2, function(x) rank(x))
data <- apply(data, 2, function(x) rescale(x, to = c(0, 100)))
shap_values <- as.matrix(read.csv("./other_data/mb_ad_shap_values_cv_data_sets.csv", check.names = FALSE))
shap_values <- as.data.frame(rescale(shap_values, to = c(0, 100), from = range(mb_data$log_intensity)))
var_diff <- apply(shap_values, 2, function(x) max(x) - min(x))
var_diff <- var_diff[order(-var_diff)]
df <- data.frame(
variable = melt(shap_values)$variable,
shap = melt(shap_values)$value,
variable_value = melt(data)$value
)top_vars <- names(var_diff)[1:20]
df <- df[df$variable %in% top_vars, ]
df$variable <- factor(df$variable, levels = rev(top_vars), labels = rev(top_vars))
p1 <- ggplot(df, aes(x = shap, y = variable)) +
geom_hline(yintercept = top_vars, linetype = "dotted", color = "grey80") +
geom_vline(xintercept = 0, color = "grey80", size = 1) +
geom_point(aes(fill = variable_value), color = "grey30", shape = 21, alpha = 0.3, size = 2, position = "auto", stroke = 0.1) +
scale_fill_gradient2(low = "#1f77b4", mid = "white", high = "#d62728", midpoint = 50, breaks = c(0, 100), labels = c("Low", "High")) +
theme_bw() +
theme(
plot.margin = ggplot2::margin(1.5, 8, 1.5, -3),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
strip.background = element_blank(),
panel.border = element_blank(),
axis.line = element_line(color = "black"),
plot.title = element_text(hjust = 0.5, size = 20),
axis.title.x = element_text(hjust = 0.5, colour = "black", size = 20),
axis.title.y = element_text(colour = "black", size = 20),
axis.text.x = element_text(colour = "black", size = 17),
axis.text.y = element_text(colour = "black", size = 16),
legend.title = element_text(size = 20),
legend.text = element_text(colour = "black", size = 16),
legend.title.align = 0.5
) +
guides(
fill = guide_colourbar("Variable\nvalue\n", ticks = FALSE, barheight = 10, barwidth = 0.5),
color = "none"
) +
xlab("SHAP value\n(variable contribution to model prediction)") +
ylab("") +
ggtitle("Melanin binding")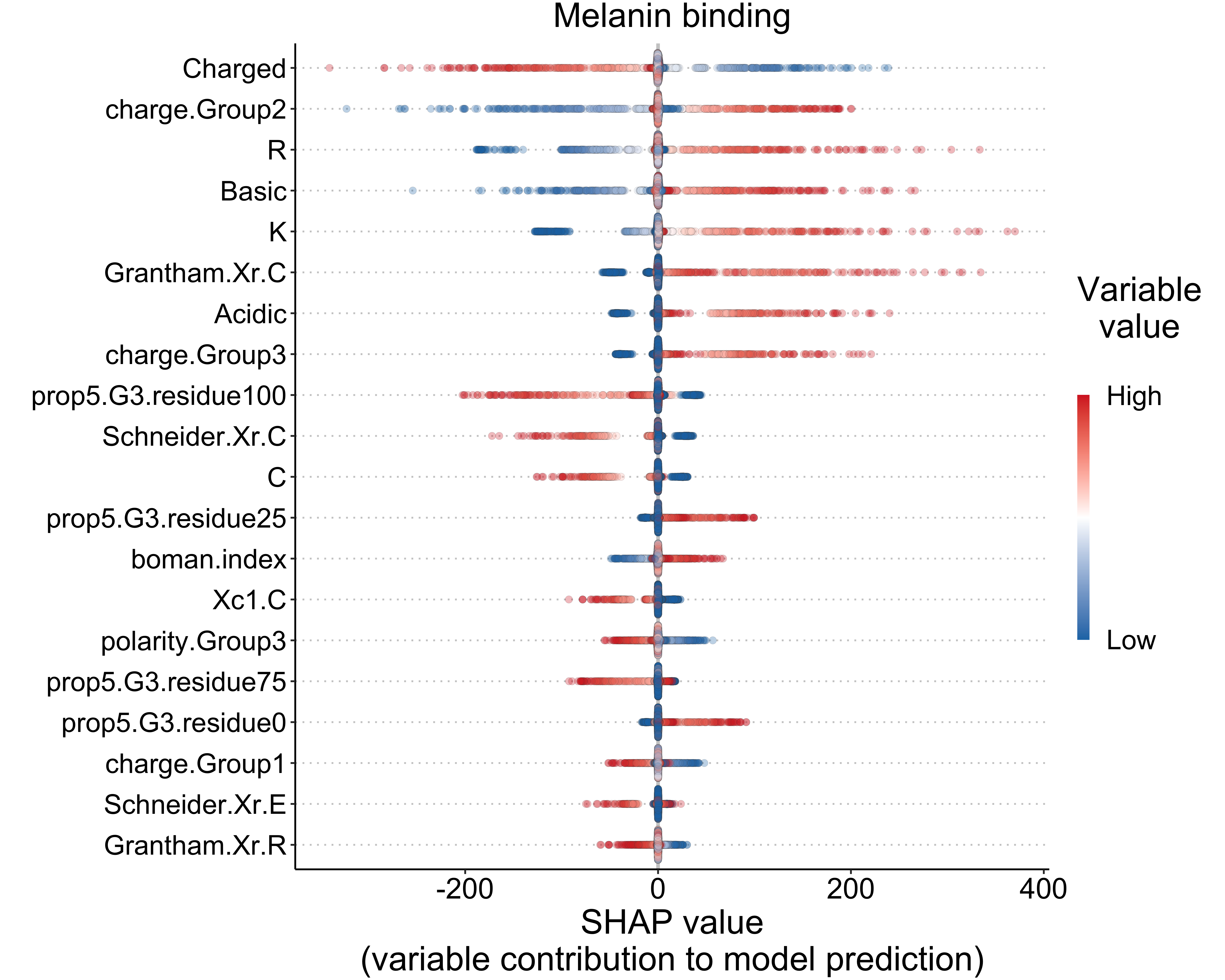
5.5.2 Cell-penetration (classification)
# Save train and test sets in csv format
load(file = "./rdata/var_reduct_cpp_train_test_splits_y_shuffled.RData")
for (i in 1:10) {
prefix <- paste0("outer_", i)
exp_dir <- paste0("/Users/renee/Downloads/cell_penetration_adversarial/", prefix)
write.csv(outer_splits[[i]]$train, file = paste0(exp_dir, "/train_set.csv"))
write.csv(outer_splits[[i]]$test, file = paste0(exp_dir, "/test_set.csv"))
}h2o.init(nthreads=-1)
dir_path = '/Users/renee/Downloads/cell_penetration_adversarial'
model_names = ['GBM_model_1671047404901_26693', 'GBM_model_1671047404901_85146', 'GBM_model_1671047404901_143415',
'GBM_model_1671047404901_201211', 'GBM_model_1671047404901_258107', 'GBM_model_1671047404901_315866',
'GBM_model_1671047404901_373347', 'GBM_model_1671047404901_432044', 'GBM_model_1671122685293_29338',
'GBM_model_1671122685293_86356']
shap_res = []
for i in range(10):
print('Iter ' + str(i + 1) + ':')
train = pd.read_csv(dir_path + '/outer_' + str(i + 1) + '/train_set.csv', index_col=0)
X_train = train.iloc[:, :-1]
test = pd.read_csv(dir_path + '/outer_' + str(i + 1) + '/test_set.csv', index_col=0)
X_test = test.iloc[:, :-1]
model = h2o.load_model(dir_path + '/outer_' + str(i + 1) + '/' + model_names[i])
def model_predict_(data):
data = pd.DataFrame(data, columns=X_train.columns)
h2o_data = h2o.H2OFrame(data)
res = model.predict(h2o_data)
res = res.as_data_frame().iloc[:,2]
return(res)
np.random.seed(1)
X_train_ = X_train.iloc[np.random.choice(X_train.shape[0], 100, replace=False), :]
explainer = shap.KernelExplainer(model_predict_, X_train_, link='identity')
shap_values = explainer.shap_values(X_test, nsamples=1000)
shap_res.append(shap_values)
h2o.remove_all()
shap_res = pd.DataFrame(np.vstack(shap_res), columns=X_train.columns)
shap_res.to_csv('./other_data/cpp_ad_shap_values_cv_data_sets.csv', index=False)load(file = "./rdata/var_reduct_cpp_train_test_splits_y_shuffled.RData")
data <- do.call(rbind, lapply(outer_splits, function(x) x$test[-ncol(cpp_data)]))
data <- apply(data, 2, function(x) rank(x))
data <- apply(data, 2, function(x) rescale(x, to = c(0, 100)))
shap_values <- read.csv("./other_data/cpp_ad_shap_values_cv_data_sets.csv", check.names = FALSE) * 100
var_diff <- apply(shap_values, 2, function(x) max(x) - min(x))
var_diff <- var_diff[order(-var_diff)]
df <- data.frame(
variable = melt(shap_values)$variable,
shap = melt(shap_values)$value,
variable_value = melt(data)$value
)top_vars <- names(var_diff)[1:11]
df <- df[df$variable %in% top_vars, ]
df$variable <- factor(df$variable, levels = rev(top_vars), labels = rev(top_vars))
p2 <- ggplot(df, aes(x = shap, y = variable)) +
geom_hline(yintercept = top_vars, linetype = "dotted", color = "grey80") +
geom_vline(xintercept = 0, color = "grey80", size = 1) +
geom_point(aes(fill = variable_value), color = "grey30", shape = 21, alpha = 0.5, size = 2, position = "auto", stroke = 0.1) +
scale_fill_gradient2(low = "#1f77b4", mid = "white", high = "#d62728", midpoint = 50, breaks = c(0, 100), labels = c("Low", "High")) +
theme_bw() +
theme(
plot.margin = ggplot2::margin(1.5, 8, 1.5, -3),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
strip.background = element_blank(),
panel.border = element_blank(),
axis.line = element_line(color = "black"),
plot.title = element_text(hjust = 0.5, size = 20),
axis.title.x = element_text(hjust = 0.5, colour = "black", size = 20),
axis.title.y = element_text(colour = "black", size = 20),
axis.text.x = element_text(colour = "black", size = 17),
axis.text.y = element_text(colour = "black", size = 16),
legend.title = element_text(size = 20),
legend.text = element_text(colour = "black", size = 16),
legend.title.align = 0.5
) +
guides(
fill = guide_colourbar("Variable\nvalue\n", ticks = FALSE, barheight = 10, barwidth = 0.5),
color = "none"
) +
xlab("SHAP value\n(variable contribution to model prediction)") +
ylab("") +
ggtitle("Cell-penetration")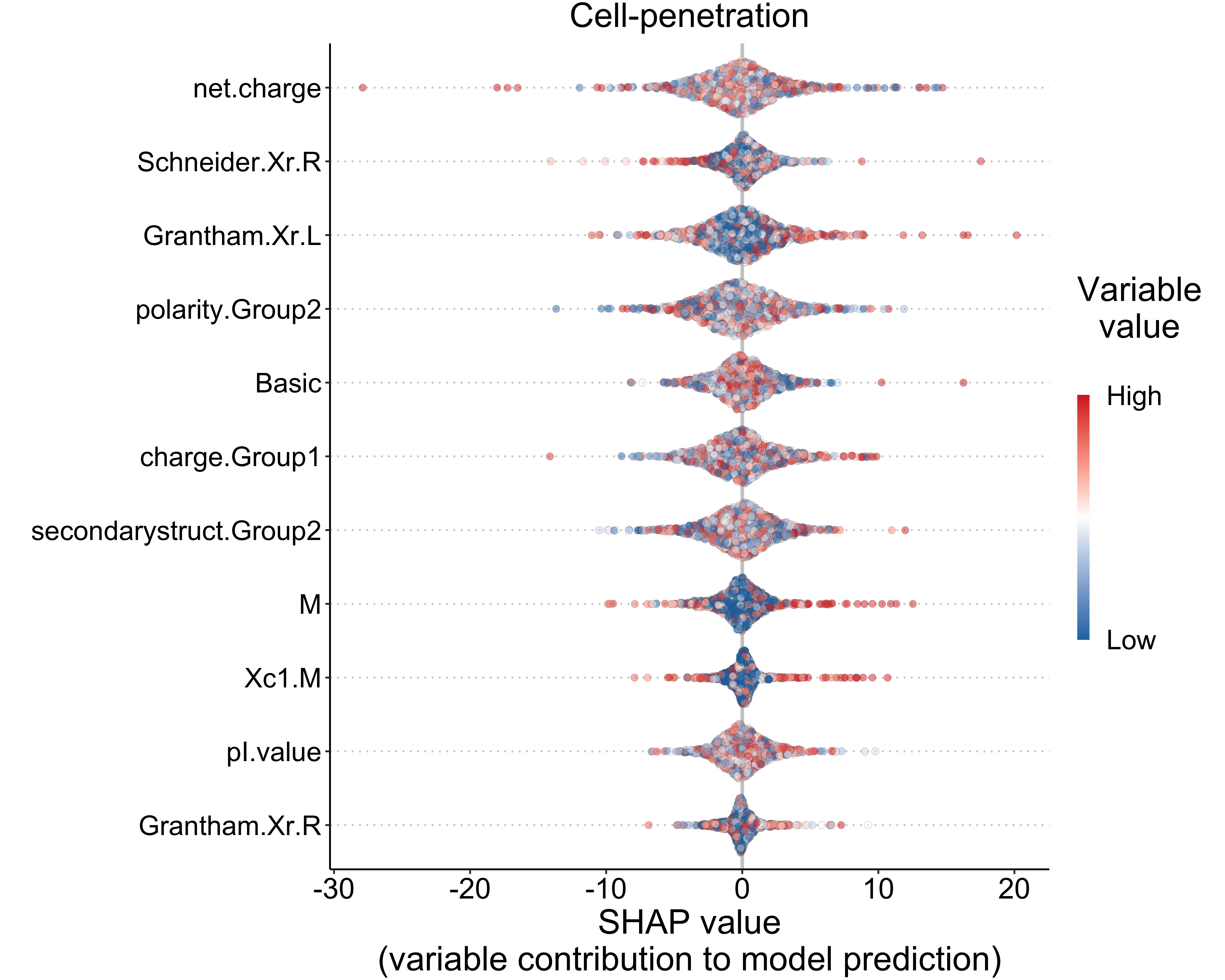
5.5.3 Toxicity (classification)
# Save train and test sets in csv format
load(file = "./rdata/var_reduct_tx_train_test_splits_y_shuffled.RData")
for (i in 1:10) {
prefix <- paste0("outer_", i)
exp_dir <- paste0("/Users/renee/Downloads/toxicity_adversarial/", prefix)
write.csv(outer_splits[[i]]$train, file = paste0(exp_dir, "/train_set.csv"))
write.csv(outer_splits[[i]]$test, file = paste0(exp_dir, "/test_set.csv"))
}h2o.init(nthreads=-1)
dir_path = '/Users/renee/Downloads/toxicity_adversarial'
model_names = ['superlearner_iter_5', 'superlearner_iter_6', 'superlearner_iter_5', 'superlearner_iter_7',
'superlearner_iter_4', 'superlearner_iter_6', 'superlearner_iter_6', 'superlearner_iter_5',
'superlearner_iter_4', 'superlearner_iter_5']
shap_res = []
for i in range(10):
print('Iter ' + str(i + 1) + ':')
train = pd.read_csv(dir_path + '/outer_' + str(i + 1) + '/train_set.csv', index_col=0)
X_train = train.iloc[:, :-1]
test = pd.read_csv(dir_path + '/outer_' + str(i + 1) + '/test_set.csv', index_col=0)
X_test = test.iloc[:, :-1]
model = h2o.load_model(dir_path + '/outer_' + str(i + 1) + '/' + model_names[i])
def model_predict_(data):
data = pd.DataFrame(data, columns=X_train.columns)
h2o_data = h2o.H2OFrame(data)
res = model.predict(h2o_data)
res = res.as_data_frame().iloc[:,2] # 0:toxic; 1:non-toxic
return(res)
np.random.seed(1)
X_train_ = X_train.iloc[np.random.choice(X_train.shape[0], 100, replace=False), :]
explainer = shap.KernelExplainer(model_predict_, X_train_, link='identity')
shap_values = explainer.shap_values(X_test, nsamples=1000)
shap_res.append(shap_values)
h2o.remove_all()
shap_res = pd.DataFrame(np.vstack(shap_res), columns=X_train.columns)
shap_res.to_csv('./other_data/tx_ad_shap_values_cv_data_sets.csv', index=False)load(file = "./rdata/var_reduct_tx_train_test_splits_y_shuffled.RData")
data <- do.call(rbind, lapply(outer_splits, function(x) x$test[-ncol(tx_data)]))
data <- apply(data, 2, function(x) rank(x))
data <- apply(data, 2, function(x) rescale(x, to = c(0, 100)))
shap_values <- read.csv("./other_data/tx_ad_shap_values_cv_data_sets.csv", check.names = FALSE) * 100
set.seed(12)
ind <- sample(1:nrow(data), 2500)
data <- data[ind, ]
shap_values <- shap_values[ind, ]
var_diff <- apply(shap_values, 2, function(x) max(x) - min(x))
var_diff <- var_diff[order(-var_diff)]
df <- data.frame(
variable = melt(shap_values)$variable,
shap = melt(shap_values)$value,
variable_value = melt(data)$value
)top_vars <- names(var_diff)[1:20]
df <- df[df$variable %in% top_vars, ]
df$variable <- factor(df$variable, levels = rev(top_vars), labels = rev(top_vars))
p3 <- ggplot(df, aes(x = shap, y = variable)) +
geom_hline(yintercept = top_vars, linetype = "dotted", color = "grey80") +
geom_vline(xintercept = 0, color = "grey80", size = 1) +
geom_point(aes(fill = variable_value), color = "grey30", shape = 21, alpha = 0.3, size = 2, position = "auto", stroke = 0.1) +
scale_fill_gradient2(low = "#1f77b4", mid = "white", high = "#d62728", midpoint = 50, breaks = c(0, 100), labels = c("Low", "High")) +
theme_bw() +
theme(
plot.margin = ggplot2::margin(1.5, 8, 1.5, -3),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
strip.background = element_blank(),
panel.border = element_blank(),
axis.line = element_line(color = "black"),
plot.title = element_text(hjust = 0.5, size = 20),
axis.title.x = element_text(hjust = 0.5, colour = "black", size = 20),
axis.title.y = element_text(colour = "black", size = 20),
axis.text.x = element_text(colour = "black", size = 17),
axis.text.y = element_text(colour = "black", size = 16),
legend.title = element_text(size = 20),
legend.text = element_text(colour = "black", size = 16),
legend.title.align = 0.5
) +
guides(
fill = guide_colourbar("Variable\nvalue\n", ticks = FALSE, barheight = 10, barwidth = 0.5),
color = "none"
) +
xlab("SHAP value\n(variable contribution to model prediction)") +
ylab("") +
ggtitle("Non-toxic")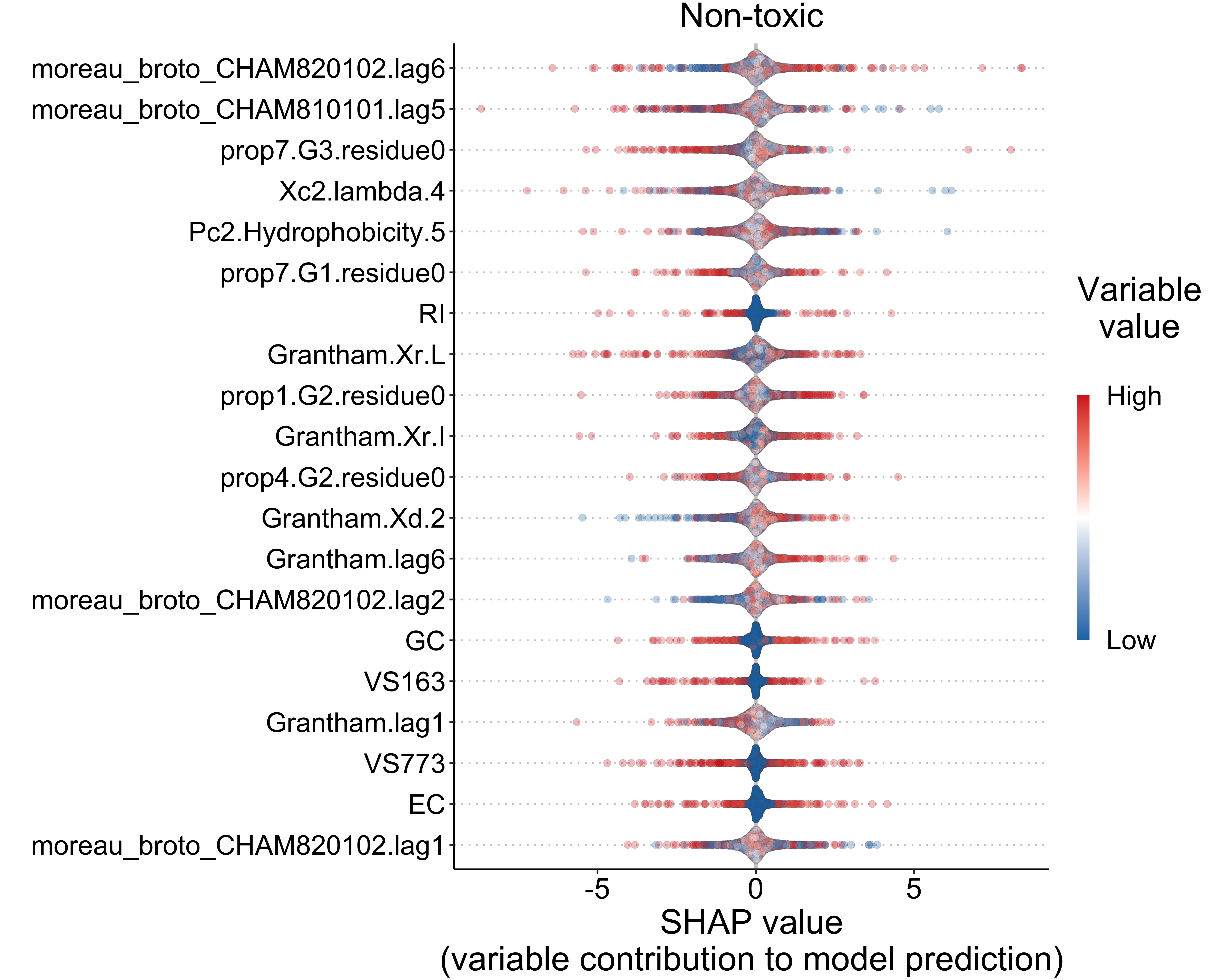
sessionInfo()## R version 4.2.2 (2022-10-31)
## Platform: x86_64-apple-darwin17.0 (64-bit)
## Running under: macOS Big Sur ... 10.16
##
## Matrix products: default
## BLAS: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRblas.0.dylib
## LAPACK: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRlapack.dylib
##
## locale:
## [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] reticulate_1.25 ggforce_0.3.4 ggplot2_3.3.6 reshape2_1.4.4
## [5] DT_0.24 digest_0.6.29 stringr_1.4.1 rlist_0.4.6.2
## [9] enrichvs_0.0.5 scales_1.2.1 mltools_0.3.5 MLmetrics_1.1.1
## [13] h2o_3.38.0.2
##
## loaded via a namespace (and not attached):
## [1] Rcpp_1.0.9 here_1.0.1 lattice_0.20-45 png_0.1-7
## [5] rprojroot_2.0.3 assertthat_0.2.1 utf8_1.2.2 R6_2.5.1
## [9] plyr_1.8.7 evaluate_0.16 highr_0.9 pillar_1.8.1
## [13] rlang_1.0.4 rstudioapi_0.14 data.table_1.14.2 jquerylib_0.1.4
## [17] R.utils_2.12.0 R.oo_1.25.0 Matrix_1.5-1 rmarkdown_2.16
## [21] styler_1.8.0 htmlwidgets_1.5.4 RCurl_1.98-1.8 polyclip_1.10-0
## [25] munsell_0.5.0 compiler_4.2.2 xfun_0.32 pkgconfig_2.0.3
## [29] htmltools_0.5.3 tidyselect_1.1.2 tibble_3.1.8 bookdown_0.28
## [33] codetools_0.2-18 fansi_1.0.3 dplyr_1.0.9 withr_2.5.0
## [37] MASS_7.3-58.1 bitops_1.0-7 R.methodsS3_1.8.2 grid_4.2.2
## [41] jsonlite_1.8.0 gtable_0.3.0 lifecycle_1.0.1 DBI_1.1.3
## [45] magrittr_2.0.3 cli_3.3.0 stringi_1.7.8 cachem_1.0.6
## [49] farver_2.1.1 bslib_0.4.0 vctrs_0.4.1 generics_0.1.3
## [53] tools_4.2.2 R.cache_0.16.0 glue_1.6.2 tweenr_2.0.1
## [57] purrr_0.3.4 crosstalk_1.2.0 fastmap_1.1.0 yaml_2.3.5
## [61] colorspace_2.0-3 knitr_1.40 sass_0.4.2session_info.show()## -----
## h2o 3.38.0.2
## matplotlib 3.6.2
## numpy 1.23.5
## pandas 1.5.2
## session_info 1.0.0
## shap 0.41.0
## -----
## Python 3.8.15 | packaged by conda-forge | (default, Nov 22 2022, 09:04:40) [Clang 14.0.6 ]
## macOS-10.16-x86_64-i386-64bit
## -----
## Session information updated at 2023-03-12 23:26