Section 5 Further characterization
5.1 Gene ontology analysis
Gene ontology (GO) enrichment analysis using GOATOOLS (Klopfenstein et al. 2018).
5.1.1 Analysis
In bash:
python ./other_data/goea.py -s "./other_data/top_200_candidates_gene_accession_1.csv" \
-p "./other_data/PlasmoDB-59_Pfalciparum3D7_GO.gaf" \
-n "./other_data/go-basic.obo" \
-o "./other_data/goea_result_1.xlsx"
python ./other_data/goea.py -s "./other_data/top_200_candidates_gene_accession_2.csv" \
-p "./other_data/PlasmoDB-59_Pfalciparum3D7_GO.gaf" \
-n "./other_data/go-basic.obo" \
-o "./other_data/goea_result_2.xlsx"
python ./other_data/goea.py -s "./other_data/top_200_candidates_gene_accession_3.csv" \
-p "./other_data/PlasmoDB-59_Pfalciparum3D7_GO.gaf" \
-n "./other_data/go-basic.obo" \
-o "./other_data/goea_result_3.xlsx"5.1.2 Plotting
In R:
library(ggplot2)
library(gridExtra)
library(shadowtext)
library(ggforce)
library(reshape)
library(cowplot)process_goea_res <- function(file_name) {
data <- read.csv(file_name, header = TRUE, fill = TRUE, quote = '\"', stringsAsFactors = FALSE)
# calculate percentage from the ratio column
data$num_genes <- apply(data, 1, function(x) as.integer(unlist(strsplit(x["ratio_in_study"], "/"))[1]))
# select for enrichment (e) data rows
data <- data[data$enrichment == "e", ]
# select for biological process (BP) data rows
res_bp <- data[data$NS == "BP", ]
res_bp$`-Log10FDR` <- -log10(res_bp$p_fdr_bh)
res_bp <- res_bp[order(res_bp$`-Log10FDR`), ]
res_bp$group <- rep("Biological process", nrow(res_bp))
# select for cellular component (CC) data rows
res_cc <- data[data$NS == "CC", ]
res_cc$`-Log10FDR` <- -log10(res_cc$p_fdr_bh)
res_cc <- res_cc[order(res_cc$`-Log10FDR`), ]
res_cc$group <- rep("Cellular component", nrow(res_cc))
# select for molecular function (MF) data rows
res_mf <- data[data$NS == "MF", ]
res_mf$`-Log10FDR` <- -log10(res_mf$p_fdr_bh)
res_mf <- res_mf[order(res_mf$`-Log10FDR`), ]
res_mf$group <- rep("Molecular function", nrow(res_mf))
ds <- do.call(rbind, list(res_mf, res_cc, res_bp))
ds$name <- factor(ds$name, levels = ds$name)
ds$group <- factor(ds$group, levels = c("Biological process", "Cellular component", "Molecular function"))
return(ds)
}ds_1 <- process_goea_res("./other_data/goea_result_1.csv")
ds_2 <- process_goea_res("./other_data/goea_result_2.csv")
ds_3 <- process_goea_res("./other_data/goea_result_3.csv")p1 <- ggplot(ds_1, aes(x = name, y = `-Log10FDR`)) +
geom_col(width = 0.05) +
geom_point(size = 5, shape = 21, color = "black", fill = "grey20") +
geom_text(aes(label = sprintf("%.0f", num_genes)), nudge_y = 0, size = 2.5, color = "white") +
coord_flip() +
ggforce::facet_col(facets = vars(group), scales = "free_y", space = "free") +
theme_bw() +
theme(
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.spacing = unit(0, "pt"),
strip.background = element_blank(),
panel.border = element_rect(colour = "black", size = 0.5),
plot.title = element_text(hjust = 0.5),
plot.margin = ggplot2::margin(5, 5, 85, 5, "pt"),
legend.text = element_text(colour = "black", ),
axis.title.x = element_text(colour = "black", ),
axis.title.y = element_text(colour = "black", ),
axis.text.x = element_text(colour = "black", ),
axis.text.y = element_text(colour = "black", )
) +
xlab("") +
ylab(expression("-Log"[10] * "FDR")) +
ggtitle("Group 1 (61 candidates)")p2 <- ggplot(ds_2, aes(x = name, y = `-Log10FDR`)) +
geom_col(width = 0.05) +
geom_point(size = 5, shape = 21, color = "black", fill = "grey20") +
geom_text(aes(label = sprintf("%.0f", num_genes)), nudge_y = 0, size = 2.5, color = "white") +
coord_flip() +
ggforce::facet_col(facets = vars(group), scales = "free_y", space = "free") +
theme_bw() +
theme(
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.spacing = unit(0, "pt"),
strip.background = element_blank(),
panel.border = element_rect(colour = "black", size = 0.5),
plot.title = element_text(hjust = 0.5),
plot.margin = ggplot2::margin(5, 0, 5, 5, "pt"),
legend.text = element_text(colour = "black", ),
axis.title.x = element_text(colour = "black", ),
axis.title.y = element_text(colour = "black", ),
axis.text.x = element_text(colour = "black", ),
axis.text.y = element_text(colour = "black", )
) +
xlab("") +
ylab(expression("-Log"[10] * "FDR")) +
ggtitle("Group 2 (83 candidates)")p3 <- ggplot(ds_3, aes(x = name, y = `-Log10FDR`)) +
geom_col(width = 0.05) +
geom_point(size = 5, shape = 21, color = "black", fill = "grey20") +
geom_text(aes(label = sprintf("%.0f", num_genes)), nudge_y = 0, size = 2.5, color = "white") +
coord_flip() +
ggforce::facet_col(facets = vars(group), scales = "free_y", space = "free") +
theme_bw() +
theme(
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.spacing = unit(0, "pt"),
strip.background = element_blank(),
panel.border = element_rect(colour = "black", size = 0.5),
plot.title = element_text(hjust = 0.5),
plot.margin = ggplot2::margin(5, 5, 5, 0, "pt"),
legend.text = element_text(colour = "black", ),
axis.title.x = element_text(colour = "black", ),
axis.title.y = element_text(colour = "black", ),
axis.text.x = element_text(colour = "black", ),
axis.text.y = element_text(colour = "black", )
) +
xlab("") +
ylab(expression("-Log"[10] * "FDR")) +
ggtitle("Group 3 (56 candidates)")Final plot
p_combined <- plot_grid(p1, p2, p3, nrow = 1, rel_widths = c(0.39, 0.34, 0.4))
p_combined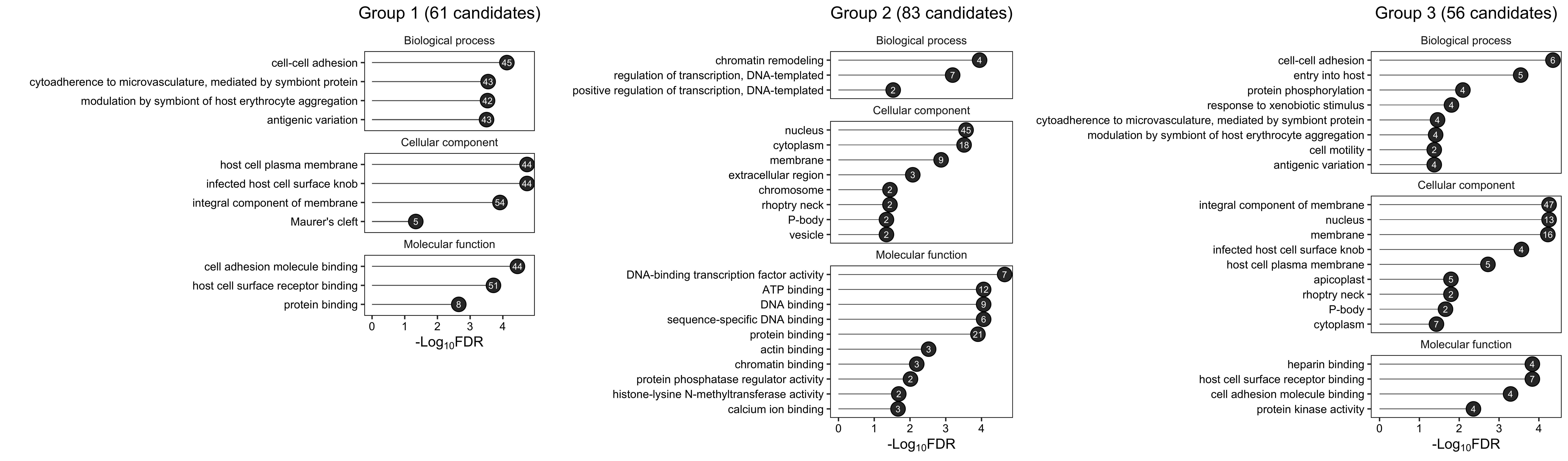
5.2 Candidate antigen characterization
Candidate antigen down-selection with essential genes (Zhang et al. 2018) and characterization using single cell RNA-seq data (Howick et al. 2019).
5.2.1 Analysis
In R:
5.2.1.1 Down-selection with essential genes
library(RMariaDB)
library(DBI)db <- dbConnect(RMariaDB::MariaDB(), user = "root", password = "", dbname = "pf_reverse_vaccinology")
get_data <- function(table_name, db) {
sql <- sqlInterpolate(db, "SELECT * FROM ?table", table = dbQuoteIdentifier(db, table_name))
return(subset(dbGetQuery(db, sql), select = -c(id)))
}
# pf3d7_essential_gene
id_map <- dbGetQuery(db, 'SELECT dbxref_1.accession AS accession, dbxref_1.dbxref_id AS transcript_id FROM dbxref AS dbxref_1
LEFT OUTER JOIN feature AS feature_1 ON dbxref_1.dbxref_id = feature_1.dbxref_id
LEFT OUTER JOIN cvterm ON feature_1.type_id = cvterm.cvterm_id
LEFT OUTER JOIN feature_relationship ON feature_1.feature_id = feature_relationship.subject_id
LEFT OUTER JOIN feature AS feature_2 ON feature_relationship.object_id = feature_2.feature_id
LEFT OUTER JOIN dbxref AS dbxref_2 ON feature_2.dbxref_id = dbxref_2.dbxref_id
LEFT OUTER JOIN organism ON feature_1.organism_id = organism.organism_id
WHERE feature_1.is_obsolete = 0 AND cvterm.name = "CDS" AND organism.species="falciparum_3D7"')
data <- get_data("pf3d7_essential_gene", db)
data <- merge(x = id_map, y = data, by = "transcript_id", all = FALSE)
accession <- data$accession
data <- subset(data, select = -c(transcript_id, accession))
rownames(data) <- accession
write.csv(data, file = "./other_data/pf_essential_genes.csv")
dbDisconnect(db)5.2.1.2 Characterization using scRNA-seq
# db = dbConnect(RMariaDB::MariaDB(), user='root', password='', dbname = 'pf_reverse_vaccinology')
#
# get_data = function(table_name, db) {
# sql = sqlInterpolate(db, 'SELECT * FROM ?table', table=dbQuoteIdentifier(db, table_name))
# return(subset(dbGetQuery(db, sql), select = -c(id)))
# }
#
# # scRNA-seq metadata
# sc_metadata = get_data("pf3d7_single_cell_coldata", db)
# rownames(sc_metadata) = sc_metadata[, 1]
# sc_metadata[, 1] = NULL
# write.csv(sc_metadata, file='./other_data/pf_sc_metadata.csv')
#
# # scRNA-seq count data
# id_map = dbGetQuery(db, 'SELECT dbxref_1.accession AS accession, dbxref_1.dbxref_id AS transcript_id FROM dbxref AS dbxref_1
# LEFT OUTER JOIN feature AS feature_1 ON dbxref_1.dbxref_id = feature_1.dbxref_id
# LEFT OUTER JOIN cvterm ON feature_1.type_id = cvterm.cvterm_id
# LEFT OUTER JOIN feature_relationship ON feature_1.feature_id = feature_relationship.subject_id
# LEFT OUTER JOIN feature AS feature_2 ON feature_relationship.object_id = feature_2.feature_id
# LEFT OUTER JOIN dbxref AS dbxref_2 ON feature_2.dbxref_id = dbxref_2.dbxref_id
# LEFT OUTER JOIN organism ON feature_1.organism_id = organism.organism_id
# WHERE feature_1.is_obsolete = 0 AND cvterm.name = "CDS" AND organism.species="falciparum_3D7"')
#
# sc_data_1 = get_data("pf3d7_single_cell_count", db)
# sc_data_2 = get_data("pf3d7_single_cell_count_2", db)
# sc_data = merge(x=sc_data_1, y=sc_data_2, by='transcript_id', all.x=TRUE)
# sc_data = merge(x=id_map, y=sc_data, by='transcript_id', all=FALSE)
# accession = sc_data$accession
# sc_data = subset(sc_data, select = -c(transcript_id, accession))
# rownames(sc_data) = accession
# write.csv(sc_data, file='./other_data/pf_sc_data.csv')
#
# dbDisconnect(db)5.2.2 Processing table
In R:
library(reshape2)
library(ggplot2)
library(ggridges)
library(rlist)
library(cowplot)
library(PupillometryR)
library(grid)
library(gridExtra)prediction <- read.csv("./data/supplementary_data_4_purf_oob_predictions.csv", row.names = 1)
prediction <- prediction[order(-prediction$oob_score_with_tree_filtering), ]
labeled_pos <- rownames(prediction)[prediction$antigen_label == 1]
top_200 <- rownames(prediction[prediction$antigen_label == 0, ])[1:200]
essential_genes <- read.csv("./other_data/pf_essential_genes.csv", header = TRUE, row.names = 1)
essential_genes <- rownames(essential_genes)[essential_genes$mis < 0.5]
essential_genes <- sapply(essential_genes, function(x) paste0(x, "-p1"))
down_selected <- top_200[top_200 %in% essential_genes]
labeled_pos <- sapply(labeled_pos, function(x) substr(x, 1, nchar(x) - 3))
top_200_ <- sapply(top_200, function(x) substr(x, 1, nchar(x) - 3))
# Downloaded from https://www.malariacellatlas.org/data-sets/, or
# https://www.malariacellatlas.org/downloads/pf-ss2-set1.zip
sc_metadata <- read.csv("~/Downloads/pf-ss2-set1/pf-ss2-set1-ss2-data.csv", row.names = 1)
# Already normalized
sc_data <- read.csv("~/Downloads/pf-ss2-set1/pf-ss2-set1-ss2-exp.csv", row.names = 1)
rownames(sc_data) <- paste0(rownames(sc_data), ".1")
sc_data_ <- sc_data[rownames(sc_data) %in% top_200_ | rownames(sc_data) %in% labeled_pos, ]
stage_order <- c(
"sporozoite (oocyst)", "sporozoite (hemolymph)", "sporozoite (salivary gland)",
"sporozoite (injected)", "sporozoite (activated)", "ring", "trophozoite", "schizont",
"gametocyte (developing)", "gametocyte (male)", "gametocyte (female)", "ookinete"
)
stage_name <- stage_orderstat_res <- list()
for (i in 1:nrow(sc_data_)) {
gene_stat_res <- c()
for (stage in stage_order) {
tmp <- as.numeric(sc_data_[i, rownames(sc_metadata[sc_metadata$STAGE_HR == stage, ])])
gene_stat_res <- c(gene_stat_res, paste0(
"Prop. cells: ", round(sum(tmp > 0) / length(tmp), 2),
"; Median: ", round(median(tmp), 2),
"; Mean: ", round(mean(tmp), 2)
))
}
stat_res <- list.append(stat_res, gene_stat_res)
}prox_w_tree_filtering <- read.csv("./other_data/proximity_values_tree_filtering.csv", check.names = FALSE)
rownames(prox_w_tree_filtering) <- colnames(prox_w_tree_filtering)
dist_w_tree_filtering <- 1 - prox_w_tree_filtering
dist_w_tree_filtering <- dist_w_tree_filtering[, names(labeled_pos)]
colnames(dist_w_tree_filtering) <- names(labeled_pos)# Prepare final table
ds <- t(data.frame(stat_res))
rownames(ds) <- sapply(rownames(sc_data_), function(x) paste0(x, "-p1"))
colnames(ds) <- stage_name
known_antigens <- sapply(labeled_pos, function(x) paste0(x, "-p1"))
# Add prediction scores
prediction <- read.csv("./data/supplementary_data_4_purf_oob_predictions.csv", row.names = 1, check.names = FALSE)
ds <- merge(
x = ds, y = prediction[, c("oob_score_with_tree_filtering"), drop = FALSE],
by = "row.names", all.x = TRUE
)
colnames(ds) <- c("Protein ID", stage_name, "Score")
# Add clustering groups
load(file = "./rdata/clustering_groups.RData")
ds$Group <- ""
ds$Group[ds$`Protein ID` %in% protein_id_1] <- "Group 1"
ds$Group[ds$`Protein ID` %in% protein_id_2] <- "Group 2"
ds$Group[ds$`Protein ID` %in% protein_id_3] <- "Group 3"
ds$Group[ds$`Protein ID` %in% known_antigens] <- "Known antigen"
ds$Group[ds$`Protein ID` %in% c(
"PF3D7_0304600.1-p1", "PF3D7_0424100.1-p1",
"PF3D7_0206900.1-p1", "PF3D7_0209000.1-p1"
)] <- "Reference antigen"
# Add gene products
gene_products <- read.csv("./other_data/pf3d7_gene_products_v59.csv")
colnames(gene_products) <- c("Protein ID", "Gene product")
ds <- merge(x = ds, y = gene_products, by = "Protein ID", all.x = TRUE)
# Add closest reference antigen and the distance
ds$`Closest known antigen` <- ""
ds$`Closest distance` <- -1
for (i in 1:nrow(ds)) {
prot <- ds$`Protein ID`[i]
tmp <- dist_w_tree_filtering[prot, ]
ds[i, "Closest known antigen"] <- names(tmp)[which.min(tmp)]
ds[i, "Closest known antigen"] <- paste0(
ds[i, "Closest known antigen"], " (",
gene_products[
gene_products$`Protein ID` == ds[
i,
"Closest known antigen"
],
"Gene product"
], ")"
)
ds[i, "Closest distance"] <- tmp[which.min(tmp)]
}
# Add essential gene information
ds$`Essential` <- "FALSE"
ds$`Essential`[ds$`Protein ID` %in% down_selected] <- "TRUE"
# Sort based on scores
ds <- ds[order(-ds$Score), ]
write.csv(ds, file = "./data/supplementary_data_7_antigen_characterization.csv", row.names = FALSE)5.2.3 Plotting
In R:
library(ggplot2)
library(gridExtra)
library(ggforce)
library(reshape)
library(stringr)data <- read.csv("./data/supplementary_data_7_antigen_characterization.csv", check.names = FALSE)
ds_0 <- data[data$Group == "Reference antigen", 1:13]
data <- data[data$Essential == TRUE, ]ds_ <- melt(ds_0, id.vars = "Protein ID")
ds_$`Protein ID` <- factor(ds_$`Protein ID`, levels = c(
"PF3D7_0304600.1-p1", "PF3D7_0424100.1-p1",
"PF3D7_0206900.1-p1", "PF3D7_0209000.1-p1"
))
ds_$prop <- sapply(ds_$value, function(x) as.numeric(str_extract_all(x, "\\d+\\.\\d+|\\d+")[[1]][1]))
ds_$median <- sapply(ds_$value, function(x) as.numeric(str_extract_all(x, "\\d+\\.\\d+|\\d+")[[1]][2]))
p0 <- ggplot(ds_, aes(x = variable, y = `Protein ID`, color = median)) +
geom_point(aes(size = prop)) +
scale_y_discrete(
limits = rev, breaks = c(
"PF3D7_0304600.1-p1", "PF3D7_0424100.1-p1",
"PF3D7_0206900.1-p1", "PF3D7_0209000.1-p1"
),
labels = c("CSP", "RH5", "MSP5", "P230")
) +
scale_color_gradient(low = "blue", high = "red", limits = c(0, 22)) +
scale_size_continuous(range = c(-1, 5), breaks = seq(0.25, 1, 0.25), limits = c(0, 1)) +
theme_bw() +
theme(
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
plot.margin = ggplot2::margin(5, 5, 0, 69, "pt"),
plot.title = element_text(hjust = 0.5),
axis.title = element_text(colour = "black"),
axis.text.x = element_blank(),
axis.ticks.x = element_blank(),
axis.text.y = element_text(colour = "black"),
rect = element_rect(fill = "transparent"),
legend.position = "none"
) +
xlab("") +
ylab("") +
ggtitle("Reference antigens")ds_1 <- data[data$Group == "Group 1", 1:13]
ds_ <- melt(ds_1, id.vars = "Protein ID")
ds_$prop <- sapply(ds_$value, function(x) as.numeric(str_extract_all(x, "\\d+\\.\\d+|\\d+")[[1]][1]))
ds_$median <- sapply(ds_$value, function(x) as.numeric(str_extract_all(x, "\\d+\\.\\d+|\\d+")[[1]][2]))
p1 <- ggplot(ds_, aes(x = variable, y = `Protein ID`, color = median)) +
geom_point(aes(size = prop)) +
scale_y_discrete(limits = rev) +
scale_color_gradient(low = "blue", high = "red", limits = c(0, 22)) +
scale_size_continuous(range = c(-1, 5), breaks = seq(0.25, 1, 0.25), limits = c(0, 1)) +
theme_bw() +
theme(
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
plot.margin = ggplot2::margin(5, 5, 0, 5, "pt"),
plot.title = element_text(hjust = 0.5),
axis.title = element_text(colour = "black"),
axis.text.x = element_blank(),
axis.ticks.x = element_blank(),
axis.text.y = element_text(colour = "black"),
rect = element_rect(fill = "transparent"),
legend.position = "none"
) +
xlab("") +
ylab("") +
ggtitle("Group 1 (2 candidates)")ds_2 <- data[data$Group == "Group 2", 1:13]
ds_ <- melt(ds_2, id.vars = "Protein ID")
ds_$prop <- sapply(ds_$value, function(x) as.numeric(str_extract_all(x, "\\d+\\.\\d+|\\d+")[[1]][1]))
ds_$median <- sapply(ds_$value, function(x) as.numeric(str_extract_all(x, "\\d+\\.\\d+|\\d+")[[1]][2]))
p2 <- ggplot(ds_, aes(x = variable, y = `Protein ID`, color = median)) +
geom_point(aes(size = prop)) +
scale_y_discrete(limits = rev) +
scale_color_gradient(low = "blue", high = "red", limits = c(0, 22)) +
scale_size_continuous(range = c(-1, 5), breaks = seq(0.25, 1, 0.25), limits = c(0, 1)) +
theme_bw() +
theme(
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
plot.title = element_text(hjust = 0.5),
plot.margin = ggplot2::margin(5, 5, 5, 5, "pt"),
axis.title = element_text(colour = "black"),
axis.text.x = element_text(colour = "black", angle = 30, vjust = 1, hjust = 1),
axis.text.y = element_text(colour = "black"),
rect = element_rect(fill = "transparent"),
legend.position = "right"
) +
guides(
colour = guide_colourbar(
title = "Median count",
title.position = "top"
),
size = guide_legend(
title = "Proportion of cells",
title.position = "top"
)
) +
xlab("") +
ylab("") +
ggtitle("Group 2 (26 candidates)")ds_3 <- data[data$Group == "Group 3", 1:13]
ds_ <- melt(ds_3, id.vars = "Protein ID")
ds_$prop <- sapply(ds_$value, function(x) as.numeric(str_extract_all(x, "\\d+\\.\\d+|\\d+")[[1]][1]))
ds_$median <- sapply(ds_$value, function(x) as.numeric(str_extract_all(x, "\\d+\\.\\d+|\\d+")[[1]][2]))
p3 <- ggplot(ds_, aes(x = variable, y = `Protein ID`, color = median)) +
geom_point(aes(size = prop)) +
scale_y_discrete(limits = rev) +
scale_color_gradient(low = "blue", high = "red", limits = c(0, 22)) +
scale_size_continuous(range = c(-1, 5), breaks = seq(0.25, 1, 0.25), limits = c(0, 1)) +
theme_bw() +
theme(
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
plot.margin = ggplot2::margin(5, 5, 5, 5, "pt"),
plot.title = element_text(hjust = 0.5),
axis.title = element_text(colour = "black"),
axis.text.x = element_text(colour = "black", angle = 30, vjust = 1, hjust = 1),
axis.text.y = element_text(colour = "black"),
rect = element_rect(fill = "transparent"),
legend.background = element_blank(),
legend.key = element_blank(),
legend.position = "none"
) +
xlab("") +
ylab("") +
ggtitle("Group 3 (14 candidates)")Final plot
p_combined <- plot_grid(plot_grid(p0, p1, p3, ncol = 1, rel_heights = c(0.2, 0.15, 0.65)),
p2,
nrow = 1, rel_widths = c(0.3, 0.4)
)
p_combined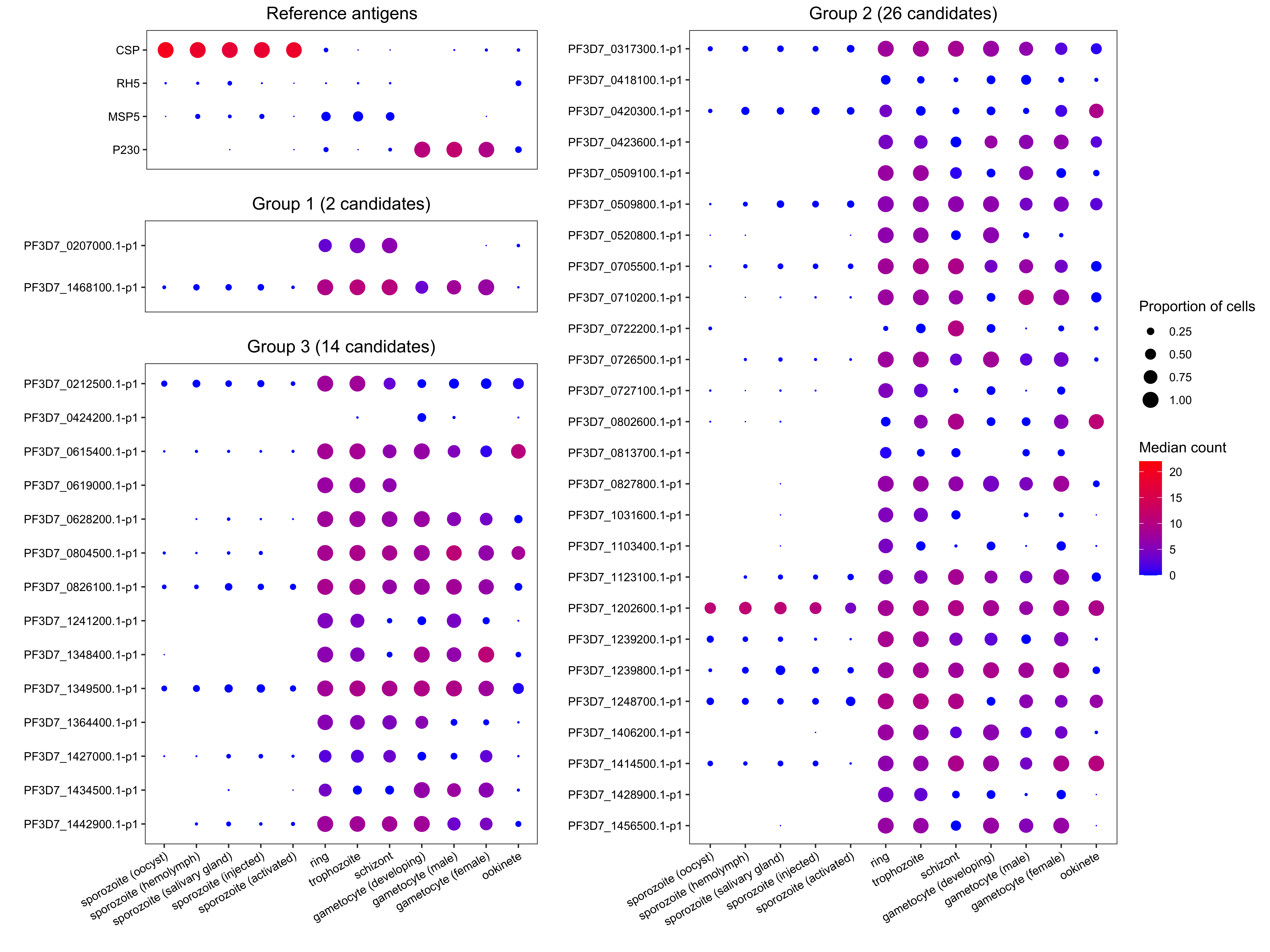
sessionInfo()## R version 4.2.3 (2023-03-15)
## Platform: x86_64-apple-darwin17.0 (64-bit)
## Running under: macOS Big Sur ... 10.16
##
## Matrix products: default
## BLAS: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRblas.0.dylib
## LAPACK: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRlapack.dylib
##
## locale:
## [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
##
## attached base packages:
## [1] grid stats graphics grDevices utils datasets methods
## [8] base
##
## other attached packages:
## [1] stringr_1.4.1 PupillometryR_0.0.4 rlang_1.1.0
## [4] dplyr_1.0.9 rlist_0.4.6.2 ggridges_0.5.3
## [7] reshape2_1.4.4 DBI_1.1.3 RMariaDB_1.2.2
## [10] cowplot_1.1.1 reshape_0.8.9 ggforce_0.3.4
## [13] shadowtext_0.1.2 gridExtra_2.3 ggplot2_3.4.2
##
## loaded via a namespace (and not attached):
## [1] Rcpp_1.0.9 assertthat_0.2.1 digest_0.6.29 utf8_1.2.2
## [5] R6_2.5.1 plyr_1.8.7 evaluate_0.16 highr_0.9
## [9] pillar_1.8.1 data.table_1.14.2 rstudioapi_0.14 jquerylib_0.1.4
## [13] R.utils_2.12.0 R.oo_1.25.0 rmarkdown_2.16 styler_1.8.0
## [17] polyclip_1.10-0 bit_4.0.4 munsell_0.5.0 compiler_4.2.3
## [21] xfun_0.32 pkgconfig_2.0.3 htmltools_0.5.3 tidyselect_1.1.2
## [25] tibble_3.1.8 bookdown_0.28 codetools_0.2-19 fansi_1.0.3
## [29] withr_2.5.0 MASS_7.3-58.2 R.methodsS3_1.8.2 jsonlite_1.8.0
## [33] gtable_0.3.0 lifecycle_1.0.3 magrittr_2.0.3 scales_1.2.1
## [37] cli_3.6.1 stringi_1.7.8 cachem_1.0.6 farver_2.1.1
## [41] bslib_0.4.0 ellipsis_0.3.2 generics_0.1.3 vctrs_0.6.2
## [45] tools_4.2.3 bit64_4.0.5 R.cache_0.16.0 glue_1.6.2
## [49] tweenr_2.0.1 purrr_0.3.4 hms_1.1.2 fastmap_1.1.0
## [53] yaml_2.3.5 colorspace_2.0-3 knitr_1.40 sass_0.4.2References
Howick, Virginia M, Andrew J C Russell, Tallulah Andrews, Haynes Heaton, Adam J Reid, Kedar Natarajan, Hellen Butungi, et al. 2019. “The Malaria Cell Atlas: Single Parasite Transcriptomes Across the Complete Plasmodium Life Cycle.” Science 365 (6455). https://doi.org/10.1126/science.aaw2619.
Klopfenstein, D V, Liangsheng Zhang, Brent S Pedersen, Fidel Ramı́rez, Alex Warwick Vesztrocy, Aurélien Naldi, Christopher J Mungall, et al. 2018. “GOATOOLS: A Python Library for Gene Ontology Analyses.” Sci Rep 8 (1): 10872. https://doi.org/10.1038/s41598-018-28948-z.
Zhang, Min, Chengqi Wang, Thomas D Otto, Jenna Oberstaller, Xiangyun Liao, Swamy R Adapa, Kenneth Udenze, et al. 2018. “Uncovering the Essential Genes of the Human Malaria Parasite Plasmodium Falciparum by Saturation Mutagenesis.” Science 360 (6388). https://doi.org/10.1126/science.aap7847.