Section 3 Model evaluation
3.1 Known antigen prediction accuracy
3.1.1 Analysis
In Python:
library(reticulate)
use_condaenv("/Users/renee/Library/r-miniconda/envs/purf/bin/python")import pandas as pd
from sklearn.ensemble import RandomForestClassifier
import numpy as np
from sklearn.impute import SimpleImputer
from sklearn.preprocessing import MinMaxScaler
from scipy.spatial import distance
from purf.pu_ensemble import PURandomForestClassifier
import pickle
import os
import re
import session_info3.1.1.1 Pv model cross-species predictions
data_ = pd.read_csv('./other_data/pv_ml_input.csv', index_col=0)
X_ = data_.iloc[:,1:]
imputer = SimpleImputer(strategy='median')
imputer.fit(X_)
data = pd.read_csv('./other_data/pf_ml_input.csv', index_col=0)
X = data.iloc[:,1:]
y = np.array(data.antigen_label)
pickle_path = './pv_0.5_purf_tree_filtering.pkl'
with open(pickle_path, 'rb') as infile:
purf = pickle.load(infile)
# Convert float type
X[X.select_dtypes(np.float64).columns] = X.select_dtypes(np.float64).astype(np.float32)
# Impute missing values
X = imputer.transform(X)
pred = purf['model'].predict_proba(X)[:,1]
res = pd.DataFrame({'protein_id': data.index, 'antigen_label': y})
res['OOB score filtered'] = pred
res.to_csv('~/Downloads/pv_single_cross_predictions.csv', index=False)3.1.1.2 Pf model cross-species predictions
data_ = pd.read_csv('./other_data/pf_ml_input.csv', index_col=0)
X_ = data_.iloc[:,1:]
imputer = SimpleImputer(strategy='median')
imputer.fit(X_)
data = pd.read_csv('./other_data/pv_ml_input.csv', index_col=0)
X = data.iloc[:,1:]
y = np.array(data.antigen_label)
pickle_path = './pf_0.5_purf_tree_filtering.pkl'
with open(pickle_path, 'rb') as infile:
purf = pickle.load(infile)
# Convert float type
X[X.select_dtypes(np.float64).columns] = X.select_dtypes(np.float64).astype(np.float32)
# Impute missing values
X = imputer.transform(X)
pred = purf['model'].predict_proba(X)[:,1]
res = pd.DataFrame({'protein_id': data.index, 'antigen_label': y})
res['OOB score filtered'] = pred
res.to_csv('~/Downloads/pf_single_cross_predictions.csv', index=False)In R:
library(DT)single_model_res <- read.csv("./other_data/pf_single_pv_single_scores.csv", check.names = FALSE)
cross_pred_res <- read.csv("./other_data/pf_single_pv_single_cross_predictions.csv", check.names = FALSE)
combined_model_res <- read.csv("./data/supplementary_data_5_pfpv_purf_oob_predictions.csv", check.names = FALSE)df <- data.frame(
"PURF model" = c("Pv single model", "Pf single model", "Pv + Pf combined model"),
"Accuracy for Pv known antigens" = c(
paste0(
nrow(single_model_res[single_model_res$antigen_label == 1 & single_model_res$species == "pv" & single_model_res$`OOB score filtered` >= 0.5, ]),
" / ", nrow(single_model_res[single_model_res$antigen_label == 1 & single_model_res$species == "pv", ]),
" = ", sprintf("%0.2f", nrow(single_model_res[single_model_res$antigen_label == 1 & single_model_res$species == "pv" & single_model_res$`OOB score filtered` >= 0.5, ]) / nrow(single_model_res[single_model_res$antigen_label == 1 & single_model_res$species == "pv", ]))
),
paste0(
nrow(cross_pred_res[cross_pred_res$antigen_label == 1 & cross_pred_res$species == "pv" & cross_pred_res$`OOB score filtered` >= 0.5, ]),
" / ", nrow(cross_pred_res[cross_pred_res$antigen_label == 1 & cross_pred_res$species == "pv", ]),
" = ", sprintf("%0.2f", nrow(cross_pred_res[cross_pred_res$antigen_label == 1 & cross_pred_res$species == "pv" & cross_pred_res$`OOB score filtered` >= 0.5, ]) / nrow(cross_pred_res[cross_pred_res$antigen_label == 1 & cross_pred_res$species == "pv", ]))
),
paste0(
nrow(combined_model_res[combined_model_res$antigen_label == 1 & combined_model_res$species == "pv" & combined_model_res$`OOB score filtered` >= 0.5, ]),
" / ", nrow(combined_model_res[combined_model_res$antigen_label == 1 & combined_model_res$species == "pv", ]),
" = ", sprintf("%0.2f", nrow(combined_model_res[combined_model_res$antigen_label == 1 & combined_model_res$species == "pv" & combined_model_res$`OOB score filtered` >= 0.5, ]) / nrow(combined_model_res[combined_model_res$antigen_label == 1 & combined_model_res$species == "pv", ]))
)
),
"Accuracy for Pf known antigens" = c(
paste0(
nrow(cross_pred_res[cross_pred_res$antigen_label == 1 & cross_pred_res$species == "pf" & cross_pred_res$`OOB score filtered` >= 0.5, ]),
" / ", nrow(cross_pred_res[cross_pred_res$antigen_label == 1 & cross_pred_res$species == "pf", ]),
" = ", sprintf("%0.2f", nrow(cross_pred_res[cross_pred_res$antigen_label == 1 & cross_pred_res$species == "pf" & cross_pred_res$`OOB score filtered` >= 0.5, ]) / nrow(cross_pred_res[cross_pred_res$antigen_label == 1 & cross_pred_res$species == "pf", ]))
),
paste0(
nrow(single_model_res[single_model_res$antigen_label == 1 & single_model_res$species == "pf" & single_model_res$`OOB score filtered` >= 0.5, ]),
" / ", nrow(single_model_res[single_model_res$antigen_label == 1 & single_model_res$species == "pf", ]),
" = ", sprintf("%0.2f", nrow(single_model_res[single_model_res$antigen_label == 1 & single_model_res$species == "pf" & single_model_res$`OOB score filtered` >= 0.5, ]) / nrow(single_model_res[single_model_res$antigen_label == 1 & single_model_res$species == "pf", ]))
),
paste0(
nrow(combined_model_res[combined_model_res$antigen_label == 1 & combined_model_res$species == "pf" & combined_model_res$`OOB score filtered` >= 0.5, ]),
" / ", nrow(combined_model_res[combined_model_res$antigen_label == 1 & combined_model_res$species == "pf", ]),
" = ", sprintf("%0.2f", nrow(combined_model_res[combined_model_res$antigen_label == 1 & combined_model_res$species == "pf" & combined_model_res$`OOB score filtered` >= 0.5, ]) / nrow(combined_model_res[combined_model_res$antigen_label == 1 & combined_model_res$species == "pf", ]))
)
),
check.names = FALSE
)
save(df, file = "./rdata/known_antigen_pred_acc.RData")load("./rdata/known_antigen_pred_acc.RData")
df %>%
datatable(rownames = FALSE)pv_antigens <- single_model_res[single_model_res$antigen_label == 1 & single_model_res$species == "pv", ]$protein_id
pv_model_pf_antigens <- cross_pred_res[cross_pred_res$antigen_label == 1 & cross_pred_res$species == "pf" & cross_pred_res$`OOB score filtered` >= 0.5, ]$protein_id
pv_model_pv_antigens <- single_model_res[single_model_res$antigen_label == 1 & single_model_res$species == "pv" & single_model_res$`OOB score filtered` >= 0.5, ]$protein_id
pf_antigens <- single_model_res[single_model_res$antigen_label == 1 & single_model_res$species == "pf", ]$protein_id
pf_model_pf_antigens <- single_model_res[single_model_res$antigen_label == 1 & single_model_res$species == "pf" & single_model_res$`OOB score filtered` >= 0.5, ]$protein_id
pf_model_pv_antigens <- cross_pred_res[cross_pred_res$antigen_label == 1 & cross_pred_res$species == "pv" & cross_pred_res$`OOB score filtered` >= 0.5, ]$protein_id
pv_intersect <- intersect(pf_model_pv_antigens, pv_model_pv_antigens)
pf_intersect <- intersect(pf_model_pf_antigens, pv_model_pf_antigens)
pv_union <- union(pf_model_pv_antigens, pv_model_pv_antigens)
pf_union <- union(pf_model_pf_antigens, pv_model_pf_antigens)
df <- data.frame(
"Set" = c("Pv model only", "Intersect", "Pf model only", "Complement"),
"Pv known antigens" = c(
paste(pv_model_pv_antigens[!pv_model_pv_antigens %in% pv_intersect], collapse = ", "),
length(pv_intersect),
paste(pf_model_pv_antigens[!pf_model_pv_antigens %in% pv_intersect], collapse = ", "),
paste(pv_antigens[!pv_antigens %in% pv_union], collapse = ", ")
),
"Pf known antigens" = c(
paste(pv_model_pf_antigens[!pv_model_pf_antigens %in% pf_intersect], collapse = ", "),
length(pf_intersect),
paste(pf_model_pf_antigens[!pf_model_pf_antigens %in% pf_intersect], collapse = ", "),
paste(pf_antigens[!pf_antigens %in% pf_union], collapse = ", ")
),
check.names = FALSE
)
save(df, file = "./rdata/known_antigen_pred_set_analysis.RData")load("./rdata/known_antigen_pred_set_analysis.RData")
df %>%
datatable(rownames = FALSE)3.2 Adversarial controls
3.2.1 Analysis
In Python:
library(reticulate)
use_condaenv("/Users/renee/Library/r-miniconda/envs/purf/bin/python")import pandas as pd
from sklearn.utils import shuffle
import numpy as np
import pickle
from sklearn.impute import SimpleImputer
from purf.pu_ensemble import PURandomForestClassifier
from sklearn.preprocessing import MinMaxScaler
from scipy.spatial import distance
from joblib import Parallel, delayed
from sklearn.ensemble._forest import _generate_unsampled_indices
import os
import re
import session_info# private function for train_purf()
def _get_ref_antigen_stats(idx, tree, X, y, ref_indices, max_samples=None):
if max_samples is None:
max_samples = y.shape[0]
oob_indices = _generate_unsampled_indices(tree.random_state, y.shape[0], max_samples)
ref_oob = [i in oob_indices for i in ref_indices]
ref_pred = list()
pred = tree.predict_proba(X[ref_indices,:], check_input=False)
ref_pred = pred[:,1]
return ref_oob, ref_pred
def train_purf(features, outcome, pos_level=0.5, purf=None, tree_filtering=False, ref_antigens=None, n_jobs=1):
features, outcome = shuffle(features, outcome, random_state=0)
# Imputation
imputer = SimpleImputer(strategy='median')
X = imputer.fit_transform(features)
X = pd.DataFrame(X, index=features.index, columns=features.columns)
y = outcome
features = X
print('There are %d positives out of %d samples before feature space weighting.' % (sum(y), len(y)))
# Feature space weighting
lab_pos = X.loc[y==1,:]
median = np.median(lab_pos, axis=0)
# Feature space weighting
lab_pos = X.loc[y==1,:]
median = np.median(lab_pos, axis=0)
scaler = MinMaxScaler(feature_range=(1,10))
dist = list()
for i in range(lab_pos.shape[0]):
dist.append(distance.euclidean(lab_pos.iloc[i, :], median))
dist = np.asarray(dist).reshape(-1, 1)
counts = np.round(scaler.fit_transform(dist))
counts = np.array(counts, dtype=np.int64)[:, 0]
X_temp = X.iloc[y==1, :]
X = X.iloc[y==0, :]
y = np.asarray([0] * X.shape[0] + [1] * (sum(counts)))
appended_data = [X]
for i in range(len(counts)):
appended_data.append(pd.concat([X_temp.iloc[[i]]] * counts[i]))
X = pd.concat(appended_data)
print('There are %d positives out of %d samples after feature space weighting.' % (sum(y), len(y)))
res = pd.DataFrame({'protein_id': X.index, 'antigen_label' : y})
if tree_filtering is True:
# get ref antigen indices
ref_index_dict = {ref:list() for ref in list(ref_antigens.values())}
for i in range(res.shape[0]):
if res['protein_id'][i] in list(ref_antigens.values()):
ref_index_dict[res['protein_id'][i]].append(res.index[i])
ref_indices = sum(ref_index_dict.values(), [])
# get OOB stats and predictions
X = X.astype('float32')
trees = purf.estimators_
idx_list = [i for i in range(len(trees))]
stats_res = Parallel(n_jobs=n_jobs)(
delayed(_get_ref_antigen_stats)(idx, trees[idx], np.array(X), y, ref_indices) for idx in idx_list)
# ref_oob data structure:
# rows represent individual trees
# column represent reference antigens
# cells indicate whether the reference antigen is in the OOB samples of the tree
ref_oob = np.array([ref_oob for ref_oob, ref_pred in stats_res])
# ref_pred data structure:
# rows represent individual trees
# column represent reference antigens
# cells indicate the prediction of the reference antigen by the tree
ref_pred = np.array([ref_pred for ref_oob, ref_pred in stats_res])
# analyze duplicated reference antigens as a group
cumsum_num_ref = np.cumsum(np.array([len(v) for k,v in ref_index_dict.items()]))
ref_oob_all = np.array([ref_oob[:, 0:cumsum_num_ref[i]].any(axis=1) if i == 0 else \
ref_oob[:, cumsum_num_ref[i - 1]:cumsum_num_ref[i]].any(axis=1) \
for i in range(len(ref_antigens))]).T
ref_pred_all = np.array([ref_pred[:, 0:cumsum_num_ref[i]].any(axis=1) if i == 0 else \
ref_pred[:, cumsum_num_ref[i - 1]:cumsum_num_ref[i]].sum(axis=1) \
for i in range(len(ref_antigens))]).T
# calculate number of reference antigens as OOB samples for each tree
oob_total = ref_oob_all.sum(axis=1)
# assign score of 1 to trees that correctly predict all OOB reference antigens; otherwise, assign 0 score
weights = np.zeros(len(trees))
# iterate through the trees and calculate the stats
for i in range(len(trees)):
oob_list = list(ref_oob_all[i,:])
pred_list = list(ref_pred_all[i,:])
if oob_total[i] == 0:
weights[i] = 0
else:
if sum(np.array(pred_list)[oob_list] != 0) == oob_total[i]:
weights[i] = 1
if tree_filtering is False:
# Training PURF
purf = PURandomForestClassifier(
n_estimators = 100000,
oob_score = True,
n_jobs = 64,
random_state = 42,
pos_level = pos_level
)
purf.fit(X, y)
else:
purf._set_oob_score_with_weights(np.array(X), y.reshape(-1,1), weights=weights)
# Storing results
res['OOB score'] = purf.oob_decision_function_[:,1]
res = features.merge(res.groupby('protein_id').mean(), left_index=True, right_on='protein_id')
res = res[['antigen_label', 'OOB score']]
if tree_filtering is False:
return (purf, res)
else:
return ({'model': purf, 'weights': weights}, res)3.2.1.1 Pv data set
ref_antigens = {'CSP': 'PVP01_0835600.1-p1', 'DBP': 'PVP01_0623800.1-p1', 'MSP1': 'PVP01_0728900.1-p1'}
data = pd.read_csv('./other_data/pv_ml_input.csv', index_col=0)
features = data.iloc[:, 1:]
outcome = np.array(data.antigen_label)
for (idx, (antigen, out)) in enumerate(zip(features.index, outcome)):
if out == 1:
if antigen in ref_antigens.values():
continue
outcome_ = outcome.copy()
outcome_[idx] = 0
(purf, res) = train_purf(features, outcome_, pos_level=0.5, tree_filtering=False)
(purf_filtered, res_filtered) = train_purf(features, outcome_, pos_level=0.5, purf=purf, tree_filtering=True, ref_antigens=ref_antigens)
res_filtered.to_csv('~/Downloads/pv_jackknife/' + antigen + '_res.csv')dir = '~/Downloads/pv_jackknife/'
files = os.listdir(dir)
for file in files:
if file.endswith('csv'):
tmp = pd.read_csv(dir + file, index_col=0)['antigen_label']
break
data_frames = [pd.read_csv(dir + file, index_col=0)['OOB score'] for file in files if file.endswith('csv')]
merged_df = pd.concat([tmp] + data_frames, join='outer', axis=1)
colnames = ['antigen_label'] + [re.match('PVP01_[0-9]+\.[0-9]-p1', file)[0] for file in files if file.endswith('csv')]
merged_df.columns = colnames
merged_df.to_csv('./other_data/pv_adversarial_controls.csv')3.2.1.2 Pv + Pf combined data set
ref_antigens = {'CSP (Pf)': 'PF3D7_0304600.1-p1', 'RH5 (Pf)': 'PF3D7_0424100.1-p1', 'MSP5 (Pf)': 'PF3D7_0206900.1-p1',
'P230 (Pf)': 'PF3D7_0209000.1-p1',
'CSP (Pv)': 'PVP01_0835600.1-p1', 'DBP (Pv)': 'PVP01_0623800.1-p1', 'MSP1 (Pv)': 'PVP01_0728900.1-p1'}
data = pd.read_csv('./data/supplementary_data_4_pfpv_ml_input.csv', index_col=0)
features = data.iloc[:, 1:]
outcome = np.array(data.antigen_label)
for (idx, (antigen, out)) in enumerate(zip(features.index, outcome)):
if out == 1:
if antigen in ref_antigens.values():
continue
outcome_ = outcome.copy()
outcome_[idx] = 0
(purf, res) = train_purf(features, outcome_, pos_level=0.5, tree_filtering=False)
(purf_filtered, res_filtered) = train_purf(features, outcome_, pos_level=0.5, purf=purf, tree_filtering=True, ref_antigens=ref_antigens)
res_filtered.to_csv('~/Downloads/pfpv_jackknife/' + antigen + '_res.csv')dir = '~/Downloads/pfpv_jackknife/'
files = os.listdir(dir)
for file in files:
if file.endswith('csv'):
tmp = pd.read_csv(dir + file, index_col=0)['antigen_label']
break
data_frames = [pd.read_csv(dir + file, index_col=0)['OOB score'] for file in files if file.endswith('csv')]
merged_df = pd.concat([tmp] + data_frames, join='outer', axis=1)
colnames = ['antigen_label'] + [re.match('PF3D7_[0-9]+\.[0-9]-p1', file)[0] if file.startswith('PF3D7') else re.match('PVP01_[0-9]+\.[0-9]-p1', file)[0] for file in files if file.endswith('csv')]
merged_df.columns = colnames
merged_df.to_csv('./other_data/pfpv_adversarial_controls.csv')3.2.2 Plotting
In R:
library(ggplot2)
library(ggdist)
library(gghalves)
library(cowplot)
library(grid)
library(ggbeeswarm)
library(ggpubr)pv_data <- read.csv("./data/supplementary_data_3_pv_purf_oob_predictions.csv", check.names = FALSE, row.names = 1)
pf_data <- read.csv("./other_data/pf_single_pv_single_scores.csv", check.names = FALSE, row.names = 1)
pfpv_data <- read.csv("./data/supplementary_data_5_pfpv_purf_oob_predictions.csv", check.names = FALSE, row.names = 1)
pv_ad_ctrl <- read.csv("./other_data/pv_adversarial_controls.csv", check.names = FALSE, row.names = 1)
pf_ad_ctrl <- read.csv("./other_data/pf_adversarial_controls.csv", check.names = FALSE, row.names = 1)
pfpv_ad_ctrl <- read.csv("./other_data/pfpv_adversarial_controls.csv", check.names = FALSE, row.names = 1)3.2.2.1 Mean differences in scores
pv_products <- read.csv("./other_data/pvp01_gene_products_v62.csv", row.names = 1)
pf_products <- read.csv("./other_data/pf3d7_gene_products_v62.csv", row.names = 1)
gene_products <- rbind(pf_products, pv_products)
calculate_known_antigen_scores <- function(validation_data, baseline_scores) {
scores <- c()
for (i in 2:ncol(validation_data)) {
known_antigen <- sort(colnames(validation_data))[i]
other_antigens <- sort(colnames(validation_data))[-c(1, i)]
scores <- c(scores, mean(validation_data[other_antigens, known_antigen] - baseline_scores[other_antigens, ]))
}
names(scores) <- sort(colnames(validation_data))[2:ncol(validation_data)]
return(scores)
}
pv_ad_ctrl_scores <- calculate_known_antigen_scores(pv_ad_ctrl, pv_data["OOB score filtered"])
pf_ad_ctrl_scores <- calculate_known_antigen_scores(pf_ad_ctrl, pf_data[rownames(pf_ad_ctrl), "OOB score filtered", drop = FALSE])
pfpv_ad_ctrl_scores <- calculate_known_antigen_scores(pfpv_ad_ctrl, pfpv_data["OOB score filtered"])
pv_ad_ctrl_res <- data.frame(
"score" = pv_ad_ctrl_scores,
"gene_product" = gene_products[names(pv_ad_ctrl_scores), "gene_product"],
row.names = names(pv_ad_ctrl_scores)
)
pv_ad_ctrl_res$source <- "Intersect"
literature <- c(
"PVP01_0102300.1-p1", "PVP01_0118900.1-p1", "PVP01_0202200.1-p1", "PVP01_0205500.1-p1",
"PVP01_0210500.1-p1", "PVP01_0304300.1-p1", "PVP01_0317900.1-p1", "PVP01_0418300.1-p1",
"PVP01_0418400.1-p1", "PVP01_0529100.1-p1", "PVP01_0532400.1-p1", "PVP01_0616000.1-p1",
"PVP01_0616100.1-p1", "PVP01_0701200.1-p1", "PVP01_0707700.1-p1", "PVP01_1026000.1-p1",
"PVP01_1030900.1-p1", "PVP01_1129100.1-p1"
)
IEDB <- c(
"PVP01_0106200.1-p1", "PVP01_0106300.1-p1", "PVP01_0215600.1-p1", "PVP01_0307400.1-p1",
"PVP01_0522300.1-p1", "PVP01_0813700.1-p1", "PVP01_0829000.1-p1", "PVP01_0835600.1-p1",
"PVP01_0923000.1-p1", "PVP01_0932400.1-p1", "PVP01_1240600.1-p1", "PVP01_1306000.1-p1",
"PVP01_1412900.1-p1", "PVP01_1453900.1-p1"
)
pv_ad_ctrl_res$source[rownames(pv_ad_ctrl_res) %in% literature] <- "Literature"
pv_ad_ctrl_res$source[rownames(pv_ad_ctrl_res) %in% IEDB] <- "IEDB"
pfpv_ad_ctrl_res <- data.frame(
"score" = pfpv_ad_ctrl_scores,
"gene_product" = gene_products[names(pfpv_ad_ctrl_scores), "gene_product"],
row.names = names(pfpv_ad_ctrl_scores)
)
pfpv_ad_ctrl_res$source <- "Intersect"
pfpv_ad_ctrl_res$source[rownames(pfpv_ad_ctrl_res) %in% literature] <- "Literature"
pfpv_ad_ctrl_res$source[rownames(pfpv_ad_ctrl_res) %in% IEDB] <- "IEDB"
write.csv(pv_ad_ctrl_res, file = "./other_data/pv_ad_ctrl_res.csv", row.names = TRUE)
write.csv(pfpv_ad_ctrl_res, file = "./other_data/pfpv_ad_ctrl_res.csv", row.names = TRUE)pv_ad_ctrl_res <- read.csv("./other_data/pv_ad_ctrl_res.csv", row.names = 1)
pfpv_ad_ctrl_res <- read.csv("./other_data/pfpv_ad_ctrl_res.csv", row.names = 1)
data_ <- data.frame(
group = factor(c(
rep(0, length(pv_ad_ctrl_scores)), rep(1, length(pf_ad_ctrl_scores)),
rep(2, length(pfpv_ad_ctrl_scores))
)),
score = c(pv_ad_ctrl_scores, pf_ad_ctrl_scores, pfpv_ad_ctrl_scores)
)
p <- ggplot(data_, aes(x = group, y = score)) +
geom_hline(yintercept = 0, color = "grey80", linetype = "dashed") +
ggdist::stat_halfeye(aes(color = group, fill = group), adjust = 0.5, width = 0.7, .width = 0, justification = -0.1, point_colour = NA, alpha = 0.9) +
geom_boxplot(aes(color = group), width = 0.1, outlier.color = NA, lwd = 0.5, show.legend = FALSE) +
gghalves::geom_half_point(aes(fill = group), side = "l", range_scale = 0.4, alpha = 0.6, shape = 21, color = "black", stroke = 0.1, size = 2) +
coord_flip() +
scale_color_manual(values = c("#03a1fc", "#984EA3", "#FF7F00")) +
scale_fill_manual(values = c("#03a1fc", "#984EA3", "#FF7F00")) +
scale_x_discrete(
breaks = c(0, 1, 2),
labels = c(
expression(paste(italic("P. vivax"), " model")),
expression(paste(italic("P. falciparum"), " model")),
"Combined model"
),
limits = rev
) +
theme_bw() +
theme(
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
axis.title.x = element_text(colour = "black"),
axis.title.y = element_blank(),
axis.text = element_text(colour = "black"),
plot.title = element_text(hjust = 0.5, colour = "black"),
plot.margin = ggplot2::margin(5, 5, 5, 5, "pt"),
legend.title = element_blank(),
legend.text = element_text(colour = "black"),
legend.position = "none"
) +
ylab("Mean difference in scores (proportion of votes)")png(
file = "./figures/Supplementary Fig 5.png",
width = 5000, height = 2000, res = 600
)
print(p)
dev.off()
pdf(file = "../supplementary_figures/Supplementary Fig 5.pdf", width = 10, height = 4)
print(p)
dev.off()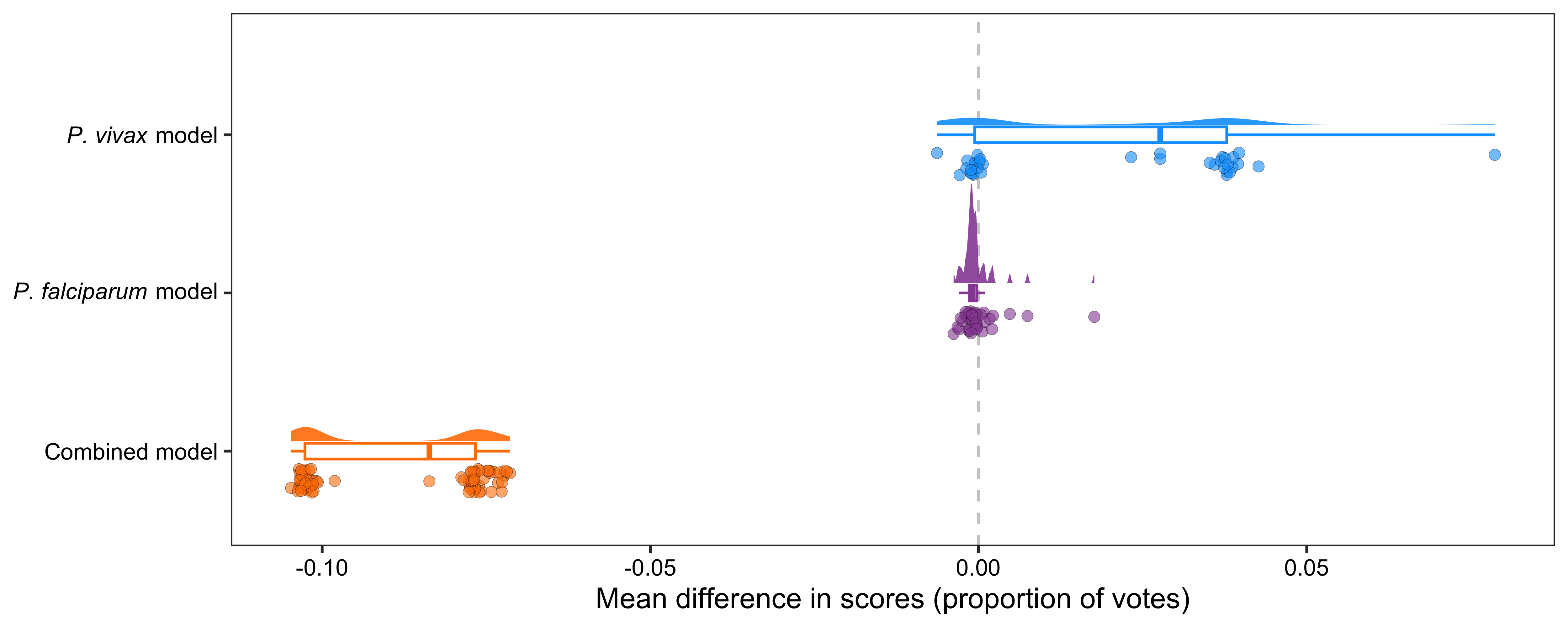
3.2.2.2 Investigation of known antigen groups
library(umap)
library(ggplot2)
library(ggrepel)
library(cowplot)pv_ad_ctrl_res <- read.csv("./other_data/pv_ad_ctrl_res.csv", row.names = 1)
# pv_ad_ctrl_res = pv_ad_ctrl_res[pv_ad_ctrl_res$source != 'Intersect', ]
pv_ad_ctrl_res$score_group <- ""
pv_ad_ctrl_res$score_group[pv_ad_ctrl_res$score < 0.01] <- "Group 1"
pv_ad_ctrl_res$score_group[pv_ad_ctrl_res$score >= 0.01] <- "Group 2"
table(pv_ad_ctrl_res$score_group, pv_ad_ctrl_res$source)##
## IEDB Intersect Literature
## Group 1 11 2 3
## Group 2 2 2 15fisher.test(pv_ad_ctrl_res$score_group, pv_ad_ctrl_res$source)##
## Fisher's Exact Test for Count Data
##
## data: pv_ad_ctrl_res$score_group and pv_ad_ctrl_res$source
## p-value = 0.0002597
## alternative hypothesis: two.sidedpfpv_ad_ctrl_res <- read.csv("./other_data/pfpv_ad_ctrl_res.csv", row.names = 1)
# pfpv_ad_ctrl_res = pfpv_ad_ctrl_res[pfpv_ad_ctrl_res$source != 'Intersect', ]
pfpv_ad_ctrl_res$score_group <- ""
pfpv_ad_ctrl_res$score_group[pfpv_ad_ctrl_res$score < -0.09] <- "Group 1"
pfpv_ad_ctrl_res$score_group[pfpv_ad_ctrl_res$score >= -0.09] <- "Group 2"
table(pfpv_ad_ctrl_res$score_group, pfpv_ad_ctrl_res$source)##
## IEDB Intersect Literature
## Group 1 12 27 2
## Group 2 1 25 16fisher.test(pfpv_ad_ctrl_res$score_group, pfpv_ad_ctrl_res$source)##
## Fisher's Exact Test for Count Data
##
## data: pfpv_ad_ctrl_res$score_group and pfpv_ad_ctrl_res$source
## p-value = 1.176e-05
## alternative hypothesis: two.sidedpfpv_ad_ctrl_res <- read.csv("./other_data/pfpv_ad_ctrl_res.csv", row.names = 1)
pfpv_ad_ctrl_res$score_group <- ""
pfpv_ad_ctrl_res$score_group[pfpv_ad_ctrl_res$score < -0.09] <- "Group 1"
pfpv_ad_ctrl_res$score_group[pfpv_ad_ctrl_res$score >= -0.09] <- "Group 2"
pfpv_ad_ctrl_res$species <- ""
pfpv_ad_ctrl_res$species[startsWith(rownames(pfpv_ad_ctrl_res), "PF3D7")] <- "Pf"
pfpv_ad_ctrl_res$species[startsWith(rownames(pfpv_ad_ctrl_res), "PVP01")] <- "Pv"
table(pfpv_ad_ctrl_res$score_group, pfpv_ad_ctrl_res$species)##
## Pf Pv
## Group 1 25 16
## Group 2 23 19fisher.test(pfpv_ad_ctrl_res$score_group, pfpv_ad_ctrl_res$species)##
## Fisher's Exact Test for Count Data
##
## data: pfpv_ad_ctrl_res$score_group and pfpv_ad_ctrl_res$species
## p-value = 0.6584
## alternative hypothesis: true odds ratio is not equal to 1
## 95 percent confidence interval:
## 0.4934941 3.3881568
## sample estimates:
## odds ratio
## 1.2867763.2.2.3 Labeled known antigen accuracies
calculate_known_antigen_accuracies <- function(validation_data, baseline_scores) {
accuracies <- c()
for (i in 2:ncol(validation_data)) {
known_antigen <- sort(colnames(validation_data))[i]
other_antigens <- sort(colnames(validation_data))[-c(1, i)]
accuracies <- c(accuracies, sum(validation_data[other_antigens, known_antigen] >= 0.5) / length(validation_data[other_antigens, known_antigen]))
}
return(accuracies)
}
pv_ad_ctrl_accuracies <- calculate_known_antigen_accuracies(pv_ad_ctrl, pv_data["OOB score filtered"])
pf_ad_ctrl_accuracies <- calculate_known_antigen_accuracies(pf_ad_ctrl, pf_data[rownames(pf_ad_ctrl), "OOB score filtered", drop = FALSE])
pfpv_ad_ctrl_accuracies <- calculate_known_antigen_accuracies(pfpv_ad_ctrl, pfpv_data["OOB score filtered"])
df <- data.frame(
"PURF model" = c("Pv single model", "Pf single model", "Pv + Pf combined model"),
"Accuracy (mean ± SD)" = c(
paste0(
sprintf("%0.2f", mean(pv_ad_ctrl_accuracies)), " ± ",
sprintf("%0.2f", sd(pv_ad_ctrl_accuracies))
),
paste0(
sprintf("%0.2f", mean(pf_ad_ctrl_accuracies)), " ± ",
sprintf("%0.2f", sd(pf_ad_ctrl_accuracies))
),
paste0(
sprintf("%0.2f", mean(pfpv_ad_ctrl_accuracies)), " ± ",
sprintf("%0.2f", sd(pfpv_ad_ctrl_accuracies))
)
),
check.names = FALSE
)
save(df, file = "./rdata/ad_ctrl_pred_accuracy.RData")load("./rdata/ad_ctrl_pred_accuracy.RData")
df %>%
datatable(rownames = FALSE)sessionInfo()## R version 4.2.3 (2023-03-15)
## Platform: x86_64-apple-darwin17.0 (64-bit)
## Running under: macOS Big Sur ... 10.16
##
## Matrix products: default
## BLAS: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRblas.0.dylib
## LAPACK: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRlapack.dylib
##
## locale:
## [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
##
## attached base packages:
## [1] grid stats graphics grDevices utils datasets methods
## [8] base
##
## other attached packages:
## [1] ggrepel_0.9.3 umap_0.2.10.0 ggpubr_0.6.0 ggbeeswarm_0.7.2
## [5] cowplot_1.1.1 gghalves_0.1.4 ggdist_3.2.1 ggplot2_3.4.2
## [9] DT_0.27 reticulate_1.28
##
## loaded via a namespace (and not attached):
## [1] Rcpp_1.0.10 here_1.0.1 lattice_0.21-8
## [4] tidyr_1.3.0 png_0.1-8 rprojroot_2.0.3
## [7] digest_0.6.31 utf8_1.2.3 RSpectra_0.16-1
## [10] R6_2.5.1 backports_1.4.1 evaluate_0.21
## [13] highr_0.10 pillar_1.9.0 rlang_1.1.1
## [16] rstudioapi_0.14 car_3.1-2 jquerylib_0.1.4
## [19] R.utils_2.12.2 R.oo_1.25.0 Matrix_1.5-4
## [22] rmarkdown_2.21 styler_1.9.1 htmlwidgets_1.6.2
## [25] munsell_0.5.0 broom_1.0.4 compiler_4.2.3
## [28] vipor_0.4.5 xfun_0.39 askpass_1.1
## [31] pkgconfig_2.0.3 htmltools_0.5.5 openssl_2.0.6
## [34] tidyselect_1.2.0 tibble_3.2.1 bookdown_0.34
## [37] codetools_0.2-19 fansi_1.0.4 dplyr_1.1.2
## [40] withr_2.5.0 R.methodsS3_1.8.2 distributional_0.3.2
## [43] jsonlite_1.8.4 gtable_0.3.3 lifecycle_1.0.3
## [46] magrittr_2.0.3 scales_1.2.1 carData_3.0-5
## [49] cli_3.6.1 cachem_1.0.8 farver_2.1.1
## [52] ggsignif_0.6.4 bslib_0.4.2 ellipsis_0.3.2
## [55] generics_0.1.3 vctrs_0.6.2 tools_4.2.3
## [58] R.cache_0.16.0 glue_1.6.2 beeswarm_0.4.0
## [61] purrr_1.0.1 crosstalk_1.2.0 abind_1.4-5
## [64] fastmap_1.1.1 yaml_2.3.7 colorspace_2.1-0
## [67] rstatix_0.7.2 knitr_1.42 sass_0.4.6session_info.show()## -----
## joblib 1.1.1
## numpy 1.19.0
## pandas 1.3.2
## purf NA
## scipy 1.8.0
## session_info 1.0.0
## sklearn 0.24.2
## -----
## Python 3.8.2 (default, Mar 26 2020, 10:45:18) [Clang 4.0.1 (tags/RELEASE_401/final)]
## macOS-10.16-x86_64-i386-64bit
## -----
## Session information updated at 2023-05-18 12:34